Bakkenolide‑IIIa ameliorates lipopolysaccharide‑induced inflammatory injury in human umbilical vein endothelial cells by upregulating LINC00294
- PMID: 33760129
- PMCID: PMC7986008
- DOI: 10.3892/mmr.2021.12016
Bakkenolide‑IIIa ameliorates lipopolysaccharide‑induced inflammatory injury in human umbilical vein endothelial cells by upregulating LINC00294
Abstract
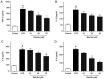
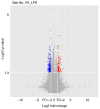
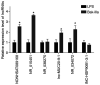
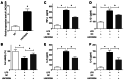
Inflammation, which causes injury to vascular endothelial cells, is one of the major factors associated with atherosclerosis (AS); therefore, inhibition of endothelial inflammation is a key step toward preventing AS. The present study aimed to investigate the effects of bakkenolide‑IIIa (Bak‑IIIa), an important active component of bakkenolides, on endothelial inflammation, as well as the mechanisms underlying such effects. Lipopolysaccharide (LPS)‑damaged human umbilical vein endothelial cells (HUVECs) were treated with Bak‑IIIa. The results of the MTT assay and enzyme‑linked immunosorbent assay indicated that Bak‑IIIa significantly alleviated survival inhibition, and decreased the levels of LPS‑induced TNF‑α, interleukin (IL)‑1β, IL‑8, and IL‑6. Furthermore, long noncoding RNA (lncRNA) microarray analyses revealed 70 differentially expressed lncRNAs (DELs) in LPS‑damaged HUVECs treated with Bak‑IIIa. lncRNA target prediction results revealed that 44 DELs had 52 cis‑targets, whereas 12 DELs covered 386 trans‑targets. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes analyses of the trans‑targets indicated that three GO terms were associated with inflammation. Therefore, 17 targets involved in these GO terms and six relevant DELs were screened out. Validation via reverse transcription‑quantitative PCR indicated that the fold change of NR_015451 (LINC00294) was the highest among the six candidates and that overexpression of LINC00294 significantly alleviated LPS‑induced survival inhibition and inflammatory damage in HUVECs. In conclusion, Bak‑IIIa ameliorated LPS‑induced inflammatory damage in HUVECs by upregulating LINC00294. Thus, Bak‑IIIa exhibited potential for preventing vascular inflammation.
Keywords: endothelial inflammation; bakkenolide‑IIIa; LINC00294.
Conflict of interest statement
All authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

