LINC01116 promotes the proliferation and invasion of glioma by regulating the microRNA‑744‑5p‑MDM2‑p53 axis
- PMID: 33760190
- PMCID: PMC7986001
- DOI: 10.3892/mmr.2021.12005
LINC01116 promotes the proliferation and invasion of glioma by regulating the microRNA‑744‑5p‑MDM2‑p53 axis
Abstract
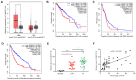
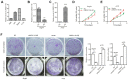
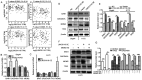
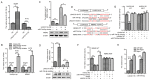
Long non‑coding RNAs (lncRNAs) have been implicated in the development and progression of tumors. However, the roles and underlying mechanisms of long intergenic non‑protein coding RNA 1116 (LINC01116), a member of the lncRNA family, in glioma progression are largely unclear. The expression of LINC01116 and microRNA (miR)‑744‑5p in glioma tissues and cells was detected by reverse transcription‑quantitative PCR. The influences of LINC01116 or miR‑744‑5p on cell proliferation and invasion were evaluated by Cell Counting Kit‑8, colony formation and Transwell assays, and western blotting was used to detect the expression of p53 pathway proteins. A dual‑luciferase reporter system was used to locate common binding sites between miR‑744‑5p and LINC01116 or the 3' untranslated region of E3 ubiquitin‑protein ligase Mdm2 (MDM2). RNA immunoprecipitation was used to determine the interactions between RNAs and proteins. Moreover, a xenograft mouse model was constructed to investigate the effects of LINC01116 in vivo, followed by a TdT‑mediated dUTP nick end labeling assay to determine the degree of apoptosis in nude mouse tumors. LINC01116 was found to be highly expressed in glioma tissues, which was associated with a malignant phenotype. LINC01116 promoted the proliferation and invasiveness of glioma cells, and inhibited the p53 pathway by preserving the expression of MDM2 mRNA via miR‑744‑5p sponging. Furthermore, a low degree of miR‑744‑5p expression was observed in glioma tissues, which was negatively associated with the expression of LINC01116. Overexpression of miR‑744‑5p inhibited the proliferation and invasiveness of glioma cells, which was rescued by LINC01116. Finally, LINC01116 knockdown inhibited tumor growth in nude mice. In conclusion, LINC01116 is aberrantly expressed and promotes the progression of glioma by regulating the miR‑744‑5p‑MDM2‑p53 pathway. In future, targeting LINC01116 may therefore be a potential therapeutic approach for patients with glioma.
Keywords: glioma; long intergenic non‑protein coding RNA 1116; microRNA‑744‑5p; MDM2; p53 pathway; competing endogenous RNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Long non‑coding RNA PCAT6 promotes the development of osteosarcoma by increasing MDM2 expression.Oncol Rep. 2020 Dec;44(6):2465-2474. doi: 10.3892/or.2020.7813. Epub 2020 Oct 16. Oncol Rep. 2020. PMID: 33125146 Free PMC article.
-
LncRNA SNHG20 promoted the proliferation of glioma cells via sponging miR-4486 to regulate the MDM2-p53 pathway.Eur Rev Med Pharmacol Sci. 2019 Jun;23(12):5323-5331. doi: 10.26355/eurrev_201906_18199. Eur Rev Med Pharmacol Sci. 2019. PMID: 31298384
-
Suppression of long non-coding RNA PCAT19 inhibits glioma cell proliferation and invasion, and increases cell apoptosis through regulation of MELK targeted by miR-142-5p.Genes Genomics. 2020 Nov;42(11):1299-1310. doi: 10.1007/s13258-020-01003-w. Epub 2020 Sep 26. Genes Genomics. 2020. PMID: 32980991
-
Promising Advances in LINC01116 Related to Cancer.Front Cell Dev Biol. 2021 Oct 14;9:736927. doi: 10.3389/fcell.2021.736927. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34722518 Free PMC article. Review.
-
The Roles Played by Long Non-Coding RNAs in Glioma Resistance.Int J Mol Sci. 2021 Jun 25;22(13):6834. doi: 10.3390/ijms22136834. Int J Mol Sci. 2021. PMID: 34202078 Free PMC article. Review.
Cited by
-
Impacts of Nutlin-3a and exercise on murine double minute 2-enriched glioma treatment.Neural Regen Res. 2025 Apr 1;20(4):1135-1152. doi: 10.4103/NRR.NRR-D-23-00875. Epub 2024 Mar 1. Neural Regen Res. 2025. PMID: 38989952 Free PMC article.
-
Promoter and enhancer RNAs regulate chromatin reorganization and activation of miR-10b/HOXD locus, and neoplastic transformation in glioma.Mol Cell. 2022 May 19;82(10):1894-1908.e5. doi: 10.1016/j.molcel.2022.03.018. Epub 2022 Apr 6. Mol Cell. 2022. PMID: 35390275 Free PMC article.
-
HOTTIP Mediated Therapy Resistance in Glioma Cells Involves Regulation of EMT-Related miR-10b.Front Oncol. 2022 Mar 24;12:873561. doi: 10.3389/fonc.2022.873561. eCollection 2022. Front Oncol. 2022. PMID: 35402278 Free PMC article.
-
Expression analysis of human glioma susceptibility gene and P53 in human glioma and its clinical significance based on bioinformatics.Ann Transl Med. 2023 Jan 31;11(2):53. doi: 10.21037/atm-22-5646. Ann Transl Med. 2023. PMID: 36819578 Free PMC article.
-
Hepatocellular Carcinoma LINC01116 Outcompetes T Cells for Linoleic Acid and Accelerates Tumor Progression.Adv Sci (Weinh). 2024 Jun;11(21):e2400676. doi: 10.1002/advs.202400676. Epub 2024 Mar 9. Adv Sci (Weinh). 2024. PMID: 38460179 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

