Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection
- PMID: 33760236
- PMCID: PMC8078597
- DOI: 10.1002/14651858.CD013705.pub2
Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection
Update in
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2025 Nov 28;11:CD013705. doi: 10.1002/14651858.CD013705.pub4. PMID: 35866452 Free PMC article. Updated.
Abstract
Background: Accurate rapid diagnostic tests for SARS-CoV-2 infection could contribute to clinical and public health strategies to manage the COVID-19 pandemic. Point-of-care antigen and molecular tests to detect current infection could increase access to testing and early confirmation of cases, and expediate clinical and public health management decisions that may reduce transmission.
Objectives: To assess the diagnostic accuracy of point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. We consider accuracy separately in symptomatic and asymptomatic population groups.
Search methods: Electronic searches of the Cochrane COVID-19 Study Register and the COVID-19 Living Evidence Database from the University of Bern (which includes daily updates from PubMed and Embase and preprints from medRxiv and bioRxiv) were undertaken on 30 Sept 2020. We checked repositories of COVID-19 publications and included independent evaluations from national reference laboratories, the Foundation for Innovative New Diagnostics and the Diagnostics Global Health website to 16 Nov 2020. We did not apply language restrictions.
Selection criteria: We included studies of people with either suspected SARS-CoV-2 infection, known SARS-CoV-2 infection or known absence of infection, or those who were being screened for infection. We included test accuracy studies of any design that evaluated commercially produced, rapid antigen or molecular tests suitable for a point-of-care setting (minimal equipment, sample preparation, and biosafety requirements, with results within two hours of sample collection). We included all reference standards that define the presence or absence of SARS-CoV-2 (including reverse transcription polymerase chain reaction (RT-PCR) tests and established diagnostic criteria).
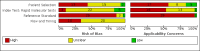
Data collection and analysis: Studies were screened independently in duplicate with disagreements resolved by discussion with a third author. Study characteristics were extracted by one author and checked by a second; extraction of study results and assessments of risk of bias and applicability (made using the QUADAS-2 tool) were undertaken independently in duplicate. We present sensitivity and specificity with 95% confidence intervals (CIs) for each test and pooled data using the bivariate model separately for antigen and molecular-based tests. We tabulated results by test manufacturer and compliance with manufacturer instructions for use and according to symptom status.
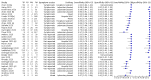
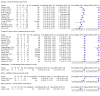
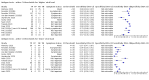
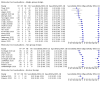
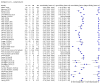
Main results: Seventy-eight study cohorts were included (described in 64 study reports, including 20 pre-prints), reporting results for 24,087 samples (7,415 with confirmed SARS-CoV-2). Studies were mainly from Europe (n = 39) or North America (n = 20), and evaluated 16 antigen and five molecular assays. We considered risk of bias to be high in 29 (50%) studies because of participant selection; in 66 (85%) because of weaknesses in the reference standard for absence of infection; and in 29 (45%) for participant flow and timing. Studies of antigen tests were of a higher methodological quality compared to studies of molecular tests, particularly regarding the risk of bias for participant selection and the index test. Characteristics of participants in 35 (45%) studies differed from those in whom the test was intended to be used and the delivery of the index test in 39 (50%) studies differed from the way in which the test was intended to be used. Nearly all studies (97%) defined the presence or absence of SARS-CoV-2 based on a single RT-PCR result, and none included participants meeting case definitions for probable COVID-19. Antigen tests Forty-eight studies reported 58 evaluations of antigen tests. Estimates of sensitivity varied considerably between studies. There were differences between symptomatic (72.0%, 95% CI 63.7% to 79.0%; 37 evaluations; 15530 samples, 4410 cases) and asymptomatic participants (58.1%, 95% CI 40.2% to 74.1%; 12 evaluations; 1581 samples, 295 cases). Average sensitivity was higher in the first week after symptom onset (78.3%, 95% CI 71.1% to 84.1%; 26 evaluations; 5769 samples, 2320 cases) than in the second week of symptoms (51.0%, 95% CI 40.8% to 61.0%; 22 evaluations; 935 samples, 692 cases). Sensitivity was high in those with cycle threshold (Ct) values on PCR ≤25 (94.5%, 95% CI 91.0% to 96.7%; 36 evaluations; 2613 cases) compared to those with Ct values >25 (40.7%, 95% CI 31.8% to 50.3%; 36 evaluations; 2632 cases). Sensitivity varied between brands. Using data from instructions for use (IFU) compliant evaluations in symptomatic participants, summary sensitivities ranged from 34.1% (95% CI 29.7% to 38.8%; Coris Bioconcept) to 88.1% (95% CI 84.2% to 91.1%; SD Biosensor STANDARD Q). Average specificities were high in symptomatic and asymptomatic participants, and for most brands (overall summary specificity 99.6%, 95% CI 99.0% to 99.8%). At 5% prevalence using data for the most sensitive assays in symptomatic people (SD Biosensor STANDARD Q and Abbott Panbio), positive predictive values (PPVs) of 84% to 90% mean that between 1 in 10 and 1 in 6 positive results will be a false positive, and between 1 in 4 and 1 in 8 cases will be missed. At 0.5% prevalence applying the same tests in asymptomatic people would result in PPVs of 11% to 28% meaning that between 7 in 10 and 9 in 10 positive results will be false positives, and between 1 in 2 and 1 in 3 cases will be missed. No studies assessed the accuracy of repeated lateral flow testing or self-testing. Rapid molecular assays Thirty studies reported 33 evaluations of five different rapid molecular tests. Sensitivities varied according to test brand. Most of the data relate to the ID NOW and Xpert Xpress assays. Using data from evaluations following the manufacturer's instructions for use, the average sensitivity of ID NOW was 73.0% (95% CI 66.8% to 78.4%) and average specificity 99.7% (95% CI 98.7% to 99.9%; 4 evaluations; 812 samples, 222 cases). For Xpert Xpress, the average sensitivity was 100% (95% CI 88.1% to 100%) and average specificity 97.2% (95% CI 89.4% to 99.3%; 2 evaluations; 100 samples, 29 cases). Insufficient data were available to investigate the effect of symptom status or time after symptom onset.
Authors' conclusions: Antigen tests vary in sensitivity. In people with signs and symptoms of COVID-19, sensitivities are highest in the first week of illness when viral loads are higher. The assays shown to meet appropriate criteria, such as WHO's priority target product profiles for COVID-19 diagnostics ('acceptable' sensitivity ≥ 80% and specificity ≥ 97%), can be considered as a replacement for laboratory-based RT-PCR when immediate decisions about patient care must be made, or where RT-PCR cannot be delivered in a timely manner. Positive predictive values suggest that confirmatory testing of those with positive results may be considered in low prevalence settings. Due to the variable sensitivity of antigen tests, people who test negative may still be infected. Evidence for testing in asymptomatic cohorts was limited. Test accuracy studies cannot adequately assess the ability of antigen tests to differentiate those who are infectious and require isolation from those who pose no risk, as there is no reference standard for infectiousness. A small number of molecular tests showed high accuracy and may be suitable alternatives to RT-PCR. However, further evaluations of the tests in settings as they are intended to be used are required to fully establish performance in practice. Several important studies in asymptomatic individuals have been reported since the close of our search and will be incorporated at the next update of this review. Comparative studies of antigen tests in their intended use settings and according to test operator (including self-testing) are required.
Copyright © 2021 The Authors. Cochrane Database of Systematic Reviews published by John Wiley & Sons, Ltd. on behalf of The Cochrane Collaboration.
Conflict of interest statement
Jonathan J Deeks: JD has published or been quoted in opinion pieces in scientific publications, and in the mainstream and social media related to diagnostic testing. JD was the statistician on the Birmingham evaluation of the Innova test which is mentioned in the discussion of the paper. There was no funding for this evaluation of the Innova test. JD is a member of the Royal Statistical Society (RSS) COVID‐19 taskforce steering group, and co‐chair of the RSS Diagnostic Test Advisory Group. He is a consultant adviser to the WHO Essential Diagnostic List. JD receives payment from the BMJ as their Chief Statistical advisor.
Jacqueline Dinnes: none known
Yemisi Takwoingi: none known
Clare Davenport: none known
Mariska MG Leeflang: none known
René Spijker: none known
Lotty Hooft: none known
Ann Van den Bruel: none known
Devy Emperador: is employed by FIND with funding from DFID and KFW. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high‐quality diagnostic tools for low‐resource settings and this is achieved by supporting both R&D and access activities for a wide range of diseases, including COVID‐19. FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Sabine Dittrich: is employed by FIND with funding from DFID and Australian Aid. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high‐quality diagnostic tools for low‐resource settings and this is achieved by supporting both R&D and access activities for a wide range of diseases, including COVID‐19. FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Ada Adriano: none known
Sophie Beese: none known
Janine Dretzke: none known
Lavinia Ferrante di Ruffano: none known
Isobel Harris: none known
Malcolm Price: none known
Sian Taylor‐Phillips: none known
Sarah Berhane: none known
Jane Cunningham: none known
Figures
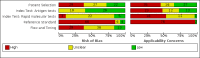



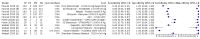
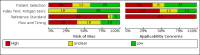
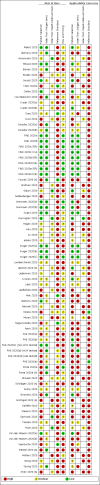
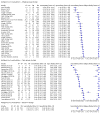
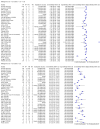



















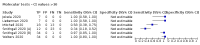
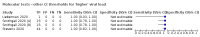
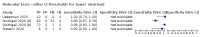
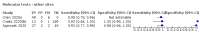

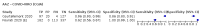

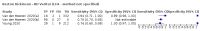
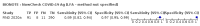

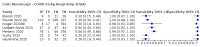
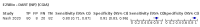
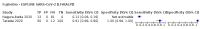
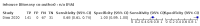




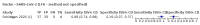

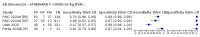

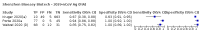


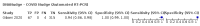
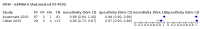
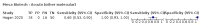




Update of
-
Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2020 Aug 26;8(8):CD013705. doi: 10.1002/14651858.CD013705. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Mar 24;3:CD013705. doi: 10.1002/14651858.CD013705.pub2. PMID: 32845525 Free PMC article. Updated.
References
References to studies included in this review
Albert 2020 {published data only}
-
- Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernández-Fuentes MÁ, et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centers. Clinical Microbiology and Infection 2020 Nov 13 [Epub ahead of print]. [DOI: 10.1016/j.cmi.2020.11.004] - DOI - PMC - PubMed
Alemany 2020 {published data only}
Assennato 2020 {published data only}
Billaud 2020 {published data only}
-
- Billaud G, Gaymard A, Lina B, Laboratoire de Virologie des HCL CNR des virus des infections respiratoires. Evaluation du Test Antigénique ABBOTT SARS-COV2 ABBOT. Lyon, France: SFM (French Society of Microbiology), 2020.
Blairon 2020 {published data only}
Broder 2020 {published data only}
Cerutti 2020 {published data only}
Chen 2020a {published data only}
Collier 2020 {published data only}
Courtellemont 2020 {published data only}
Cradic 2020(a) {published data only}
Cradic 2020(b) {published data only}
Diao 2020 {published data only}
-
- Diao B, Wen K, Chen J, Liu Y, Yuan Z, Han C, et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv [Preprint] 10 March 2020:1-13. [DOI: 10.1101/2020.03.07.20032524] - DOI
Dust 2020 {published data only}
Fenollar 2020(a) {published data only}
Fenollar 2020(b) {published data only}
FIND 2020a {published data only}
-
- Foundation for Innovative New Diagnostics (FIND). Evaluation of Bionote, Inc. NowCheck COVID-19 Ag Test - External Report. Switzerland: FIND, 2020.
FIND 2020b {published data only}
-
- Foundation for Innovative New Diagnostics (FIND). Evaluation of Abbott Panbio COVID-19 Ag Rapid Test Device - External Report. Switzerland: FIND, 2020.
FIND 2020c (BR) {published data only}
-
- Foundation for Innovative New Diagnostics (FIND). Evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test - External Report. Switzerland: FIND, 2020.
FIND 2020c (CH) {published data only}
-
- Foundation for Innovative New Diagnostics (FIND). Evaluation of SD Biosensor, Inc. STANDARD Q COVID-19 Ag Test - External Report. Switzerland: FIND, 2020.
FIND 2020d (BR) {published data only}
-
- Foundation for Innovative New Diagnostics (FIND). Evaluation of SD Biosensor, Inc. STANDARD F COVID-19 Ag FIA - External Report. Switzerland: FIND, 2020.
FIND 2020d (DE) {published data only}
-
- Foundation for Innovative New Diagnostics (FIND). Evaluation of SD Biosensor, Inc. STANDARD F COVID-19 Ag FIA - External Report. Switzerland: FIND, 2020.
FIND 2020e (BR) {published data only}
-
- Foundation for Innovative New Diagnostics (FIND). Evaluation of RapiGEN Inc. BIOCREDIT COVID-19 Ag - External Report. Switzerland: FIND, 2020.
FIND 2020e (DE) {published data only}
-
- Foundation for Innovative New Diagnostics (FIND). Evaluation of RapiGEN Inc. BIOCREDIT COVID-19 Ag - External Report. Switzerland: FIND, 2020.
Fourati 2020 [A] {published data only}
-
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020.
Fourati 2020 [B] {published data only}
-
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020.
Fourati 2020 [C] {published data only}
-
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020.
Fourati 2020 [D] {published data only}
-
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020.
Fourati 2020 [E] {published data only}
-
- Fourati S, Audureau E, Chevaliez S, Pawlotsky JM. Évaluation de la performance diagnostique des tests rapides d’orientation diagnostique antigéniques COVID-19. France: AP-HP Hopitaux universitaires Henri-Mondor, 2020.
Ghofrani 2020 {published data only}
-
- Ghofrani M, Casas MT, Pelz RK, Kroll C, Blum N, Foster SD. Performance characteristics of the ID NOW COVID-19 assay: a regional health care system experience. medRxiv [Preprint] 2020. [DOI: ]
Gibani 2020 {published data only}
Goldenberger 2020 {published data only}
Gremmels 2020(a) {published data only}
Gremmels 2020(b) {published data only}
Gupta 2020 {published data only}
-
- Gupta A, Khurana S, Das R, Srigyan D, Singh A, Mittal A, et al. Rapid chromatographic immunoassay-based evaluation of COVID-19: a cross-sectional, diagnostic test accuracy study & its implications for COVID-19 management in India. Indian Journal of Medical Research 2020 Oct 31 [Epub ahead of print]. [DOI: 10.4103/ijmr.IJMR_3305_20] - DOI - PMC - PubMed
Harrington 2020 {published data only}
-
- Harrington A, Cox B, Snowdon J, Bakst J, Ley E, Grajales P, et al. Comparison of Abbott ID NOW and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. Journal of Clinical Microbiology 2020;58(8):e00798-20. [DOI: 10.1128/JCM.00798-20.] - DOI - PMC - PubMed
Hogan 2020 {published data only}
Hou 2020 {published data only}
Jin 2020 {published data only}
-
- Jin R, Pettengill MA, Hartnett NL, Auerbach HE, Peiper SC, Wang Z. Commercial severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) molecular assays: superior analytical sensitivity of cobas SARS-CoV-2 relative to NxTAG Cov Extended Panel and ID NOW COVID-19 test. Archives of Pathology and Laboratory Medicine 2020;144(11):1303-10. - PubMed
Jokela 2020 {published data only}
Kruger 2020(a) {published data only}
-
- Krüger LJ, Gaeddert M, Köppel L, Brümmer LE, Gottschalk C, Miranda IB, et al. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ]
Kruger 2020(b) {published data only}
-
- Krüger LJ, Gaeddert M, Köppel L, Brümmer LE, Gottschalk C, Miranda IB, et al. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ]
Kruger 2020(c) {published data only}
-
- Krüger LJ, Gaeddert M, Köppel L, Brümmer LE, Gottschalk C, Miranda IB, et al. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ]
Lambert‐Niclot 2020 {published data only}
-
- Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso AM, et al. Evaluation of a rapid diagnostic assay for detection of SARS CoV-2 antigen in nasopharyngeal swab. Journal of Clinical Microbiology 2020;58(8):e00977-20. [DOI: 10.1128/JCM.00977-20] - DOI - PMC - PubMed
Lephart 2020 [A] {published data only}
-
- Lephart PR, Bachman M, LeBar W, McClellan S, Barron K, Schroeder L, et al. Comparative study of four SARS-CoV-2 nucleic acid amplification test (NAAT) platforms demonstrates that ID NOW performance is impaired substantially by patient and specimen type. bioRxiv [Preprint] 2020. [DOI: ] - PMC - PubMed
Lephart 2020 [B] {published data only}
-
- Lephart PR, Bachman M, LeBar W, McClellan S, Barron K, Schroeder L, et al. Comparative study of four SARS-CoV-2 nucleic acid amplification test (NAAT) platforms demonstrates that ID NOW performance is impaired substantially by patient and specimen type. bioRxiv [Preprint] 2020. [DOI: ] - PMC - PubMed
Lieberman 2020 {published data only}
-
- Lieberman JA, Pepper G, Naccache SN, Huang ML, Jerome KR, Greninger AL. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. Journal of Clinical Microbiology 2020;58(8):e00821-20. [DOI: 10.1128/JCM.00821-20] - DOI - PMC - PubMed
Linares 2020 {published data only}
Liotti 2020 {published data only}
-
- Liotti FM, Menchinelli G, Lalle E, Palucci I, Marchetti S, Colavita F, et al. Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clinical Microbiology and Infection 2020 Sep 23 [Epub ahead of print]. [DOI: 10.1016/j.cmi.2020.09.030] - DOI - PMC - PubMed
Loeffelholz 2020 {published data only}
Mak 2020 {published data only}
Mertens 2020 {published data only}
Mitchell 2020 {published data only}
Moore 2020 {published data only}
-
- Moore NM, Li H, Schejbal D, Lindsley J, Hayden M. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCOV RT-PCR assay for the qualitative detection of SARS-CoV-2 from upper respiratory tract specimens. medRxiv [Preprint] 2020:1-22. [DOI: 10.1101/2020.05.02.20088740] - DOI - PMC - PubMed
Moran 2020 {published data only}
Nagura‐Ikeda 2020 {published data only}
-
- Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. Journal of Clinical Microbiology 2020;58(9):e01438-20. [DOI: 10.1128/JCM.01438-20] - DOI - PMC - PubMed
Nash 2020 {published data only}
-
- Nash B, Badea A, Reddy A, Bosch M, Salcedo N, Gomez AR, et al. The impact of high frequency rapid viral antigen screening on COVID-19 spread and outcomes: a validation and modeling study. medRxiv [Preprint] 2020. [DOI: ]
PHE 2020(a) {published data only}
-
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020.
PHE 2020(b) {published data only}
-
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020.
PHE 2020(c) [non‐HCW tested] {published data only}
-
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020.
PHE 2020(d) [HCW tested] {published data only}
-
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020.
PHE 2020(d) [Lab tested] {published data only}
-
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020.
PHE 2020(e) {published data only}
-
- Public Health England (PHE). Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: rapid evaluation of lateral flow viral antigen detection devices (LFDs) for mass community testing. Public Health England, 2020.
Porte 2020a {published data only}
-
- Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, et al. Evaluation of novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. papers.ssrn.com/abstract=3569871 [Preprint] 14 April 2020;(dx.doi.org/10.2139/ssrn.3569871):1-23. [DOI: ] - PMC - PubMed
Porte 2020b [A] {published data only}
-
- Porte L, Legarraga P, Iruretagoyena M, Vollrath V, Pizarro G, Munita JM, et al. Rapid SARS-CoV-2 antigen detection by immunofluorescence – a new tool to detect infectivity. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.10.04.20206466] - DOI
Porte 2020b [B] {published data only}
-
- Porte L, Legarraga P, Iruretagoyena M, Vollrath V, Pizarro G, Munita JM, et al. Rapid SARS-CoV-2 antigen detection by immunofluorescence – a new tool to detect infectivity. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.10.04.20206466] - DOI
Rhoads 2020 {published data only}
-
- Rhoads DD, Cherian SS, Roman K, Stempak LM, Schmotzer CL, Sadri N. Comparison of Abbott ID NOW, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. Journal of Clinical Microbiology 2020;58(8):e00760-20. [DOI: 10.1128/JCM.00760-20] - DOI - PMC - PubMed
Schildgen 2020 [A] {published data only}
Schildgen 2020 [B] {published data only}
Schildgen 2020 [C] {published data only}
Scohy 2020 {published data only}
Shrestha 2020 {published data only}
-
- Shrestha B, Neupane AK, Pant S, Shrestha A, Bastola, A. Sensitivity and specificity of lateral flow antigen test kits for COVID-19 in asymptomatic population of quarantine centre of Province 3. Kathmandu University Medical Journal 2020;18(70):36-9. - PubMed
Smithgall 2020 [A] {published data only}
Smithgall 2020 [B] {published data only}
SoRelle 2020 {published data only}
-
- SoRelle Jeffrey, Mahimmainathan Lenin, McCormick-Baw Clare, Cavuoti Dominick, Lee Franceca, Bararia Anjali, et al. Evaluation of symptomatic patient saliva as a sample type for the Abbott ID NOW COVID-19 assay. medRxiv [Preprint] 2020. [DOI: ]
Stevens 2020 {published data only}
Szymczak 2020 {published data only}
Takeda 2020 {published data only}
-
- Takeda Y, Mori M, Omi K. SARS-CoV-2 qRT-PCR Ct value distribution in Japan and possible utility of rapid antigen testing kit. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.06.16.20131243] - DOI
Thwe 2020 {published data only}
-
- Thwe PM, Ren P. How many are we missing with ID NOW COVID-19 assay using direct nasopharyngeal swabs? Findings from a mid-sized academic hospital clinical microbiology laboratory. Diagnostic Microbiology and Infectious Disease 2020;98(2):115123. [DOI: 10.1016/j.diagmicrobio.2020.115123] - DOI - PMC - PubMed
Van der Moeren 2020(a) {published data only}
-
- Van der Moeren N, Zwart VF, Lodder EB, Van den Bijllaardt W, Van Esch HR, Stohr JJ, et al. Performance evaluation of a SARS-CoV-2 rapid antigen test: test performance in the community in the Netherlands. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.10.19.20215202] - DOI
Van der Moeren 2020(b) {published data only}
-
- Van der Moeren N, Zwart VF, Lodder EB, Van den Bijllaardt W, Van Esch HR, Stohr JJ, et al. Performance evaluation of a SARS-CoV-2 rapid antigen test: test performance in the community in the Netherlands. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.10.19.20215202] - DOI
Veyrenche 2020 {published data only}
Weitzel 2020 [A] {published data only}
-
- Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Porte L, et al. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Medicine and Infectious Diseases 2020 Dec 2 [Epub ahead of print]:101942. [DOI: 10.1016/j.tmaid.2020.101942] - DOI - PMC - PubMed
Weitzel 2020 [B] {published data only}
Weitzel 2020 [C] {published data only}
Weitzel 2020 [D] {published data only}
Wolters 2020 {published data only}
-
- Wolters F, Van de Bovenkamp J, Van den Bosch B, Van den Brink S, Broeders M, Chung NH, et al. Multi-center evaluation of Cepheid Xpert(R) Xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. Journal of Clinical Virology 2020;128:104426. [DOI: 10.1016/j.jcv.2020.104426] - DOI - PMC - PubMed
Wong 2020 {published data only}
Young 2020 {published data only}
-
- Young S, Taylor S, Cammarata C, Roger-Dalbert C, Montano A, Griego-Fullbright C, et al. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS Antigen point-of-care test. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.09.01.20185777] - DOI - PMC - PubMed
-
- Young S, Taylor SN, Cammarata CL, Varnado KG, Roger-Dalbert C, Montano A, et al. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS Antigen point-of-care test. Journal of Clinical Microbiology 2020. [DOI: 10.1128/JCM.02338-20] - DOI - PMC - PubMed
Zhen 2020 [A] {published data only}
References to studies excluded from this review
Ai 2020 {published data only}
-
- Ai JW, Zhang HC, Xu T, Wu J, Zhu M, Yu YQ, et al. Optimizing diagnostic strategy for novel coronavirus pneumonia, a multi-center study in Eastern China. medRxiv [Preprint] 17 February 2020:1-18. [DOI: 10.1101/2020.02.13.20022673] - DOI
Anahtar 2020 {published data only}
-
- Anahtar MN, McGrath GE, Rabe BA, Tanner NA, White BA, Lennerz JK, et al. Clinical assessment and validation of a rapid and sensitive SARS-CoV-2 test using reverse-transcription loop-mediated isothermal amplification. medRxiv [Preprint] 18 May 2020:1-22. [DOI: 10.1101/2020.05.12.20095638] - DOI - PMC - PubMed
Ar Gouilh 2020 {published data only}
-
- Ar Gouilh M, Cassier R, Maille E, Schanen C, Rocque L-M, Vabret Astrid. An easy, reliable and rapid SARS-CoV2 RT-LAMP based test for Point-of-Care and diagnostic lab. medRxiv [Preprint] 2020. [DOI: ]
Arizti‐Sanz 2020 {published data only}
Arumugam 2020 {published data only}
-
- Arumugam A, Faron ML, Yu P, Markham C, Wong S. A rapid COVID-19 RT-PCR detection assay for low resource settings. bioRxiv [Preprint] 30 April 2020:1-13. [DOI: 10.1101/2020.04.29.069591] - DOI
Avetyan 2020 {published data only}
Azhar 2020 {published data only}
Azzi 2020 {published data only}
Baek 2020 {published data only}
Barra 2020 {published data only}
Basu 2020 {published data only}
-
- Basu A, Zinger T, Inglima K, Woo KM, Atie O, Yurasits L, et al. Performance of Abbott ID NOW COVID-19 rapid nucleic acid amplification test in nasopharyngeal swabs transported in viral media and dry nasal swabs, in a New York City academic institution. Journal of Clinical Microbiology 2020;58(8):e01136-20. [DOI: 10.1128/JCM.01136-20] - DOI - PMC - PubMed
Behrmann 2020 {published data only}
-
- Behrmann O, Bachmann I, Spiegel M, Schramm M, El Wahed AA, Dobler G, et al. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (exo-IQ). Clinical Chemistry 8 May 2020 [Epub ahead of print]:hvaa116. [DOI: 10.1093/clinchem/hvaa116] - DOI - PMC - PubMed
Bokelmann 2020 {published data only}
Bordi 2020 {published data only}
Brandsma 2020 {published data only}
Broughton 2020 {published data only}
-
- Broughton JP, Deng X, Yu G, Fasching CL, Singh J, Streithorst J, et al. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. medRxiv [Preprint] 27 March 2020:1-28. [DOI: 10.1101/2020.03.06.20032334] - DOI
Bull 2020 {published data only}
Bulterys 2020 {published data only}
Callahan 2020a {published data only}
-
- Callahan CJ, Lee R, Zulauf K, Tamburello L, Smith KP, Previtera J, et al. Open development and clinical validation of multiple 3D-printed sample-collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. medRxiv [Preprint] 7 May 2020:1-16. [EMBASE: 10.1101/2020.04.14.20065094] - PMC - PubMed
-
- Callahan CJ, Lee R, Zulauf KE, Tamburello L, Smith KP, Previtera J, et al. Open development and clinical validation of multiple 3D-printed nasopharyngeal collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. Journal of Clinical Microbiology 2020;58(8):e00876-20. [DOI: 10.1128/JCM.00876-20] - DOI - PMC - PubMed
Callahan 2020b {published data only}
-
- Callahan C, Lee R, Lee G, Zulauf K E, Kirby J E, Arnaout R. Nasal-swab testing misses patients with low SARS-CoV-2 viral loads. medRxiv [Preprint] 2020. [DOI: ]
Chandler‐Brown 2020 {published data only}
-
- Chandler-Brown D, Bueno AM, Atay O, Tsao DS. A highly scalable and rapidly deployable RNA extraction-free COVID-19 assay by quantitative Sanger sequencing. medRxiv [Preprint] 10 April 2020:1-15. [DOI: 10.1101/2020.04.07.029199] - DOI
Chen 2020b {published data only}
Chow 2020 {published data only}
CNR 2020 {published data only}
-
- Centre National de Référence des virus des infections respiratoires. Evaluation des performances analytiques du test VitaPCR™ SARS-CoV-2 Assay, BIOSYNEX. Lyon, France: SFM (French Society of Microbiology), 2020.
CNR 2020a {published data only}
-
- Centre National de Référence des virus des infections respiratoires. Résultats d’évaluation de la performance en analytique pour la détection du SARS-CoV-2 dans le cadre de l’épidémie de COVID-19 comparaison avec la technique de référence du CNR IPP. Lyon, France: SFM (French Society of Microbiology), 2020.
Colson 2020 {published data only}
Comar 2020 {published data only}
-
- Comar M, Brumat M, Concas MP, Argentini G, Bianco A, Bicego L, et al. COVID-19 experience: first Italian survey on healthcare staff members from a Mother-Child Research hospital using combined molecular and rapid immunoassays test. medRxiv [Preprint] 22 April 2020:1-12. [DOI: 10.1101/2020.04.19.20071563] - DOI
Comer 2020 {published data only}
-
- Comer SW, Fisk D. An extended laboratory validation study and comparative performance evaluation of the Abbott ID NOW COVID-19 Assay in a Coastal California tertiary care medical center. medRxiv [Preprint] 2020. [DOI: ]
Crone 2020 {published data only}
-
- Crone MA, Priestman M, Ciechonska M, Jensen K, Sharp DJ, Randell P, et al. A new role for biofoundries in rapid prototyping, development, and validation of automated clinical diagnostic tests for SARS-CoV-2. medRxiv [Preprint] 12 May 2020:1-31. [DOI: 10.1101/2020.05.02.20088344] - DOI
Curti 2020 {published data only}
-
- Curti L, Pereyra-Bonnet F, Gimenez CA. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv [Preprint] 2 March 2020:1-10. [DOI: 10.1101/2020.02.29.971127] - DOI
Davda 2020 {published data only}
-
- Davda JN, Frank K, Prakash S, Purohit G, Vijayashankar DP, Vedagiri D, et al. An inexpensive RT-PCR endpoint diagnostic assay for SARS-CoV-2 using nested PCR: direct assessment of detection efficiency of RT-qPCR tests and suitability for surveillance. bioRxiv [Preprint] 2020. [DOI: ]
Ding 2020a {published data only}
-
- Ding X, Yin K, Li Z, Liu C. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv [Preprint] 21 March 2020:1-19. [DOI: 10.1101/2020.03.19.998724] - DOI
Ding 2020b {published data only}
Dohla 2020 {published data only}
Dong 2020 {published data only}
El‐Tholoth 2020 {published data only}
-
- El-Tholoth M, Bau HH, Song J. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. chemRxiv [Preprint] 2020;19:19. [DOI: 10.26434/chemrxiv.11860137] - DOI
Farfan 2020 {published data only}
FIND 2020f {published data only}
-
- FIND. FIND Evaluation of Coris BioConcept COVID-19 Ag Respi-Strip - External Report. Switzerland: FIND, 2020.
Fowler 2020 {published data only}
-
- Fowler VL, Armson B, Gonzales JL, Wise EL, Howson EL, Vincent-Mistiaen Z, et al. A reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 within nasopharyngeal and oropharyngeal swabs at Hampshire Hospitals NHS Foundation Trust. medRxiv [Preprint] 2020. [DOI: ]
Francis 2020 {published data only}
-
- Francis R, Le Bideau M, Jardot P, Grimaldier C, Raoult D, Khalil JY, et al. High speed large scale automated isolation of SARS-CoV-2 from clinical samples using miniaturized co-culture coupled with high content screening. bioRxiv [Preprint] 19 May 2020:1-23. [DOI: 10.1101/2020.05.14.097295] - DOI - PMC - PubMed
Freire‐Paspuel 2020a {published data only}
-
- Freire-Paspuel B, Vega-Marino P, Velez A, Cruz M, Bereguiain MA. High sensitivity CDC EUA SARS-CoV-2 kit-based End Point-PCR assay. medRxiv [Preprint] 18 May 2020:1-7. [DOI: 10.1101/2020.05.11.20098590] - DOI
Freire‐Paspuel 2020b {published data only}
-
- Freire-Paspuel B, Vega-Marino P, Velez A, Castillo P, Cruz M, Garcia-Bereguiain MA. Evaluation of nCoV-QS (MiCo BioMed) for RT-qPCR detection of SARS-CoV-2 from nasopharyngeal samples using CDC FDA EUA qPCR kit as a gold standard: An example of the need of validation studies. Journal of Clinical Virology 2020;128:104454. [DOI: 10.1016/j.jcv.2020.104454] - DOI - PMC - PubMed
Ganguli 2020 {published data only}
Giamarellos‐Bourboulis 2020 {published data only}
Gonzalez‐Gonzalez 2020a {published data only}
-
- Gonzalez-Gonzalez E, Lara-Mayorga IM, Rodriguez-Sanchez IP, Yee-de Leon F, Garcia-Rubio A, Garciamendez-Mijares CE, et al. Scaling diagnostics in times of COVID-19: rapid prototyping of 3D-printed water circulators for loop-mediated isothermal amplification (LAMP) and detection of SARS-CoV-2 virus. medRxiv [Preprint] 19 June 2020:1-39. [DOI: 10.1101/2020.04.09.20058651] - DOI
Gonzalez‐Gonzalez 2020b {published data only}
Grant 2020 {published data only}
-
- Grant PR, Turner MA, Shin GY, Nastouli E, Levett LJ. Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. bioRxiv [Preprint] 9 April 2020:1-6. [DOI: ]
Hass 2020 {published data only}
Herrera 2020 {published data only}
-
- Herrera V, Hsu V, Adewale A, Hendrix T, Johnson L, Kuhlman J, et al. Testing of healthcare workers exposed to COVID19 with rapid antigen detection. medRxiv [Preprint] 2020. [DOI: ]
Hirotsu 2020 {published data only}
-
- Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. International Journal of Infectious Diseases 2020;99:397-402. - PMC - PubMed
Hogan 2020a {published data only}
Howson 2020 {published data only}
-
- Howson E, Kidd S, Sawyer J, Cassar C, Cross D, Lewis T, et al. Preliminary optimisation of a simplified sample preparation method to permit direct detection of SARS-CoV-2 within saliva samples using reverse-transcription loop-mediated isothermal amplification (RT-LAMP). medRxiv [Preprint] 2020. [DOI: ] - PMC - PubMed
Hu 2020 {published data only}
Huang 2020 {published data only}
Huang 2021 {published data only}
James 2020 {published data only}
-
- James P, Stoddart D, Harrington ED, Beaulaurier J, Ly L, Reid SW, et al. LamPORE: rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv [Preprint] 2020. [DOI: ]
Jiang 2020 {published data only}
-
- Jiang M, Pan W, Arastehfar A, Fang W, ling L, Fang H, et al. Development and validation of a rapid single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. medRxiv [Preprint] 27 March 2020:1-12. [DOI: 10.1101/2020.03.15.20036376] - DOI - PMC - PubMed
Joung 2020 {published data only}
-
- Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang MW, et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv [Preprint] 8 May 2020:1-21. [DOI: 10.1101/2020.05.04.20091231] - DOI
Joung 2020a {published data only}
Kalikiri 2020 {published data only}
-
- Kalikiri MK, Hasan M, Mirza F, Xaba T, Tang P, Lorenz S. High-throughput extraction of SARS-CoV-2 RNA from nasopharyngeal swabs using solid-phase reverse immobilization beads. medRxiv [Preprint] 11 April 2020:1-5. [DOI: 10.1101/2020.04.08.20055731] - DOI
Kashiwagi 2020 {published data only}
Kim 2019 {published data only}
Kim 2020 {published data only}
-
- Kim Y, Yaseen AB, Kishi JY, Hong F, Saka SK, Sheng K, et al. Single-strand RPA for rapid and sensitive detection of SARS-CoV-2 RNA. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.08.17.20177006]
Konrad 2020 {published data only}
Kurstjens 2020 {published data only}
Kyosei 2020 {published data only}
Lalli 2020 {published data only}
-
- Lalli MA, Chen X, Langmade SJ, Fronick CC, Sawyer CS, Burcea LC, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva with colorimetric LAMP. medRxiv [Preprint] 11 May 2020:1-25. [DOI: 10.1101/2020.05.07.20093542] - DOI
Lamb 2020 {published data only}
Landry 2020 {published data only}
Lee 2020 {published data only}
-
- Lee JY, Best N, McAuley J, Porter JL, Seemann T, Schultz MB, et al. Validation of a single-step, single-tube reverse transcription-loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. bioRxiv [Preprint] 30 April 2020. [DOI: 10.1101/2020.04.28.067363] - DOI - PMC - PubMed
Le Hingrat 2020 {published data only}
-
- Le Hingrat Q, Visseaux B, Laouenan C, Tubiana S, Bouadma L, Yazdanpanah Y, et al. SARS-CoV-2 N-antigenemia: a new alternative to nucleic acid amplification techniques. medRxiv [Preprint] 2020.
Li 2020 {published data only}
Lin 2020 {published data only}
Liotti 2020a {published data only}
Lowe 2020 {published data only}
Lu 2020 {published data only}
Lu 2020a {published data only}
Lubke 2020 {published data only}
Mahari 2020 {published data only}
-
- Mahari S, Roberts A, Shahdeo D, Gandhi S. eCovSens-Ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCOVID-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv [Preprint] 11 May 2020:1-20. [DOI: 10.1101/2020.04.24.059204] - DOI
Marais 2020 {published data only}
Marzinotto 2020 {published data only}
McCormick‐Baw 2020 {published data only}
-
- McCormick-Baw C, Morgan K, Gaffney D, Cazares Y, Jaworski K, Byrd A, et al. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. Journal of Clinical Microbiology 2020;58(8):e01109-20. [DOI: 10.1128/JCM.01109-20] - DOI - PMC - PubMed
McDonald 2020 {published data only}
McRae 2020 {published data only}
Mei 2020 {published data only}
Meyerson 2020 {published data only}
-
- Meyerson NR, Yang Q, Clark SK, Paige CL, Fattor WT, Gilchrist AR, et al. A community-deployable SARS-CoV-2 screening test using raw saliva with 45 minutes sample-to-results turnaround. medRxiv [Preprint] 2020. [DOI: ]
Michel 2020 {published data only}
Mlcochova 2020 {published data only}
Mohon 2020 {published data only}
Moses 2020 {published data only}
-
- Moses SE, Warren C, Robinson P, Curtis J, Asquith S, Holme J, et al. Endpoint PCR Detection of Sars-CoV-2 RNA. medRxiv [Preprint] 2020. [DOI: ]
Mostafa 2020 {published data only}
Muraoka 2020 {published data only}
-
- Muraoka M, Tanoi Y, Tada T, Mizukoshi M, Kawaguchi O. Quickly and simply detection for coronavirus including SARS-CoV-2 on the mobile real-time PCR device and without RNA Extraction. medRxiv [Preprint] 2020. [DOI: ]
Nachtigall 2020 {published data only}
-
- Nachtigall FM, Pereira A, Trofymchuk OS, Santos LS. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nature Biotechnology 2020;38(10):1168-73. - PubMed
Newman 2020 {published data only}
Noerz 2020 {published data only}
Ogawa 2020 {published data only}
Osterdahl 2020 {published data only}
Paden 2020 {published data only}
Patchsung 2020 {published data only}
-
- Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P, et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nature Biomedical Engineering 2020;4(12):1140-9. - PubMed
Pellanda 2020 {published data only}
-
- Pellanda LC, Wendland EM, McBride AJ, Tovo-Rodrigues L, Ferreira MR, Dellagostin OA, et al. Sensitivity and specificity of a rapid test for assessment of exposure to SARS-CoV-2 in a community-based setting in Brazil. medRxiv [Preprint] 10 May 2020:1-10. [DOI: 10.1101/2020.05.06.20093476] - DOI
Peto 2020 {published data only}
Pfefferle 2020 {published data only}
Pollock 2020a {published data only}
Qian 2020 {published data only}
Rabe 2020 {published data only}
Rauch 2020 {published data only}
-
- Rauch JN, Valois E, Ponce-Rojas JC, Aralis Z, Lach RS, Zappa F, et al. CRISPR-based and RT-qPCR surveillance of SARS-CoV-2 in asymptomatic individuals uncovers a shift in viral prevalence among a university population. medRxiv [Preprint] 2020. [DOI: ]
Rodel 2020 {published data only}
Rodriguez‐Manzano 2020 {published data only}
Seo 2020 {published data only}
-
- Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020;14(4):5135-42. - PubMed
Shirato 2020 {published data only}
Singh 2020a {published data only}
Singh 2020b {published data only}
Smyrlaki 2020 {published data only}
St Hilaire 2020 {published data only}
Tan 2020 {published data only}
-
- Tan X, Lin C, Zhang J, Khaing OM, Fan X. Rapid and quantitative detection of COVID-19 markers in micro-liter sized samples. bioRxiv [Preprint] 22 April 2020:1-17. [DOI: 10.1101/2020.04.20.052233] - DOI
Tanida 2020 {published data only}
Tibbetts 2020 {published data only}
-
- Tibbetts R, Callahan K, Rofoo K, Zarbo RJ, Samuel L. Comparison of the NeuMoDX, Diasorin Simplexa, Cepheid and Roche CDC SARS-CoV 2 EUA assays using nasopharyngeal/nasal swabs in universal transport media (UTM) and sputum and tracheal aspirates. bioRxiv [Preprint] 2020. [DOI: ]
Tran 2020 {published data only}
-
- Tran DH, Cuong HQ, Tran HT, Le UP, Do HD, Kui LM, et al. A comparative study of isothermal nucleic acid amplification methods for SARS-CoV-2 detection at point of care. bioRxiv [Preprint] 2020. [DOI: ]
Visseaux 2020 {published data only}
-
- Visseaux B, Le Hingrat Q, Collin G, Bouzid D, Lebourgeois S, Le Pluart D, et al. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. Journal of Clinical Microbiology 2020;58(8):e00630-20. [DOI: 10.1128/JCM.00630-20] - DOI - PMC - PubMed
Wang 2020a {published data only}
Wang 2020b {published data only}
Wang 2020c {published data only}
Wee 2020 {published data only}
Wu 2020 {published data only}
Xue 2020 {published data only}
Yan 2020 {published data only}
Yang 2020b {published data only}
-
- Yang W, Dang X, Wang Q, Xu M, Zhao Q, Zhou Y, et al. Rapid detection of SARS-CoV-2 using reverse transcription RT-LAMP method. medRxiv [Preprint] 3 March 2020:1-25. [DOI: 10.1101/2020.03.02.20030130] - DOI
Yu 2020a {published data only}
Yu 2020b {published data only}
-
- Yu L, Wu S, Hao X, Li X, Liu X, Ye S, et al. Rapid colorimetric detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform: iLACO. medRxiv [Preprint] 24 February 2020:1-19. [DOI: 10.1101/2020.02.20.20025874] - DOI - PMC - PubMed
Yu 2020c {published data only}
-
- Yu S, Nimse SB, Kim J, Song KS, Kim T. Development of a lateral flow strip membrane assay for rapid and sensitive detection of the SARS-CoV-2. Analytical Chemistry 2020;92(20):14139-44. - PubMed
Zamecnik 2020 {published data only}
Zeng 2020 {published data only}
Zhang 2020 {published data only}
-
- Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv [Preprint] 29 February 2020:1-14. [DOI: 10.1101/2020.02.26.20028373] - DOI
Zhao 2020 {published data only}
-
- Zhao Z, Cui H, Song W, Ru X, Zhou W, Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. bioRxiv [Preprint] 27 February 2020:1-18. [DOI: 10.1101/2020.02.22.961268] - DOI
Zhu 2020 {published data only}
Additional references
Arevalo‐Rodriguez 2020
Bossuyt 2015
Boyce 2018
Carter 2020
CDC 2020
-
- Centers for Disease Control and Prevention (CDC). Interim Guidance for Antigen Testing for SARS-CoV-2. Available from: www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines... 2020.
Cevik 2021
Chen 2020
Cheng 2020
Corman 2020
-
- Corman V, Bleicker T, Brünink S, Drosten C, Landt O, Koopmans M, et al. Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR - protocol and preliminary evaluation as of Jan 13, 2020. Available from www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e... 2020.
Covidence [Computer program]
-
- Veritas Health Innovation Covidence. Version accessed 27 April 2020. Melbourne, Australia: Veritas Health Innovation. Available at covidence.org.
Crozier 2021
-
- Crozier A, Rajan S, Buchan I, McKee M. Put to the test: use of rapid testing technologies for COVID-19. BMJ 2021;372:n208. - PubMed
Deeks 2020a
-
- Deeks JJ, Raffle AE. Lateral flow tests cannot rule out SARS-CoV-2 infection. BMJ 2020;371:m4787. - PubMed
Deeks 2020b
-
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, Spijker R, et al. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews 2020, Issue 4. Art. No: CD013596. [DOI: 10.1002/14651858.CD013596] - DOI
Deeks 2020c
Ferguson 2020
FIND 2020
-
- FIND. SARS-COV-2 Diagnostic pipeline. www.finddx.org/covid-19/pipeline/ (accessed 5 January 2021).
Green 2020
-
- Green K, Graziadio S, Turner P, Fanshawe T, Allen J, on behalf of the Oxford COVID-19 Evidence Service Team Centre. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. Available at www.cebm.net/oxford-covid-19/ 7 April 2020.
Hadgu 1999
Healy 2020
Jaafar 2020
-
- Jaafar R, Aherfi S, Wurtz N, Grimaldier C, Van Hoang T, Colson P, et al. Correlation between 3790 quantitative polymerase chain reaction–positives samples and positive cell cultures, including 1941 severe acute respiratory syndrome coronavirus 2 isolates. Clinical Infectious Diseases 2020:ciaa1491. [DOI: ] - PMC - PubMed
Kozel 2017
Kretzschmar 2020
Kucirka 2020
Landier 2018
-
- Landier J, Haohankhunnatham W, Das S, Konghahong K, Christensen P, Raksuansak J, et al. Operational performance of a Plasmodium falciparum ultrasensitive rapid diagnostic test for detection of asymptomatic infections in Eastern Myanmar. Journal of Clinical Microbiology 2018;56(8):e00565-18. [DOI: 10.1128/JCM.00565-18] - DOI - PMC - PubMed
Larremore 2020
Lee 2021
-
- Lee LY, Rozmanowski S, Pang M, Charlett A, Anderson C, Hughes GJ, et al. An observational study of SARS-CoV-2 infectivity by viral load and demographic factors and the utility lateral flow devices to prevent transmission. modmedmicro.nsms.ox.ac.uk/wp-content/uploads/2021/01/infectivity_manuscr... (accessed before March 2021).
Marks 2021
Mayers 2020
-
- Mayers C, Baker K, Government Office for Science. Impact of false-positives and false-negatives in the UK’s COVID-19 RT-PCR testing programme. Available from assets.publishing.service.gov.uk/government/uploads/system/uploads/attac... 2020.
McInnes 2018
McInnes 2020
Meyerowitz 2020
Moher 2009
Nabavi 2021
-
- Nabavi N, Dobson J. Testing asymptomatic individuals for SARS-CoV-2—known unknowns. blogs.bmj.com/bmj/2021/02/19/testing-asymptomatic-individuals-for-sars-c... 2021.
ONS 2020
-
- Office for National Statistics. Coronavirus (COVID-19) Infection Survey, UK: 21 August 2020. Available from: ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsan... 2020.
Pai 2012
Pilarowski 2021
Pollock 2020
-
- Pollock NR, Jacobs JR, Tran K, Cranston A, Smith S, O’Kane C, et al. Performance and implementation evaluation of the Abbott BinaxNOW Rapid Antigen Test in a high-throughput drive-through community testing site in Massachusetts. medRxiv [Preprint] 2021. [DOI: 10.1101/2021.01.09.21249499] - DOI - PMC - PubMed
Reitsma 2005
Review Manager 2020 [Computer program]
-
- The Cochrane Collaboration Review Manager 5 (RevMan 5). Version 5.4. Copenhagen: The Cochrane Collaboration, 2020.
Riley 2020
-
- Riley S, Ainslie KE, Eales O, Walters CE, Wang H, Atchison C, et al. Transient dynamics of SARS-CoV-2 as England exited national lockdown. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.08.05.20169078] - DOI
Salameh 2020
Singanayagam 2020
Stata [Computer program]
-
- Stata. Version 15. College Station, TX, USA: StataCorp, 2017. Available at www.stata.com.
Stegeman 2020
Struyf 2021
-
- Struyf T, Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19. Cochrane Database of Systematic Reviews 2021, Issue 2. Art. No: CD013665. [DOI: 10.1002/14651858.CD013665.pub2] - DOI - PMC - PubMed
Takwoingi 2017
University of Liverpool 2020
-
- University of Liverpool. Liverpool COVID-19 community testing pilot. Interim evaluation report. Available from liverpool.ac.uk/media/livacuk/coronavirus/Liverpool,Community,Testing,Pi... 2020.
Vogels 2020
Whiting 2011
-
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. - PubMed
WHO 2018
-
- World Health Organization (WHO). Diagnostic assessment: in vitro diagnostic medical devices (IVDs) used for the detection of high-risk human papillomavirus (HPV) genotypes in cervical cancer screening. Licence: CC BY-NC-SA 3.0 IGO. Available at apps.who.int/iris/handle/10665/272282 2018.
WHO 2020a
-
- World Health Organization. WHO information notice for IVD users: nucleic acid testing (NAT) technologies that use real-time polymerase chain reaction (RT-PCR) for detection of SARS-CoV-2. Available from who.int/news/item/14-12-2020-who-information-notice-for-ivd-users 2020.
WHO 2020b
-
- World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays. Interim guidance 11 September 2020. Available from: apps.who.int/iris/bitstream/handle/10665/334253/WHO-2019-nCoV-Antigen_De... 2020.
WHO 2020c
-
- World Health Organization. COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.1.0. Available from who.int/publications/m/item/covid-19-target-product-profiles-for-priorit... 2020.
WHO 2020d
-
- World Health Organization. Laboratory testing of 2019 novel coronavirus (2019-nCoV) in suspected human cases: interim guidance. Available from www.who.int/publications/i/item/laboratory-testing-for-2019-novel-corona... 2020.
WHO 2020e
-
- Global surveillance for COVID-19 caused by human infection with COVID-19 virus: interim guidance 20 March 2020. Available from apps.who.int/iris/bitstream/handle/10665/331506/WHO-2019-nCoV-Surveillan... 2020.
References to other published versions of this review
Dinnes 2020
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

