Glycoproteomic Sample Processing, LC-MS, and Data Analysis Using GlycReSoft
- PMID: 33761173
- PMCID: PMC8349174
- DOI: 10.1002/cpz1.84
Glycoproteomic Sample Processing, LC-MS, and Data Analysis Using GlycReSoft
Erratum in
-
Group Correction Statement (Data Availability Statements).Curr Protoc. 2022 Aug;2(8):e552. doi: 10.1002/cpz1.552. Curr Protoc. 2022. PMID: 36005902 Free PMC article. No abstract available.
-
Group Correction Statement (Conflict of Interest Statements).Curr Protoc. 2022 Aug;2(8):e551. doi: 10.1002/cpz1.551. Curr Protoc. 2022. PMID: 36005903 Free PMC article. No abstract available.
Abstract
Identification of N- and O-glycosylation on specific sites of proteins, along with glycan structural information, is necessary to determine the roles glycoproteins play in normal and pathologic cellular functions. Because such glycosylation is macro- and micro-heterogeneous and alters the dissociation behavior of glycopeptides, specific sample preparation, mass spectrometry, and data analysis techniques are required. Advanced tandem mass spectrometry-based glycoproteomics coupled with powerful data mining algorithms are key elements for characterization of protein glycosylation. This article includes the detailed, streamlined sample preparation method for liquid chromatography-mass spectrometry data acquisition and subsequent bioinformatics-based data annotation using the publicly available GlycReSoft program for highly efficient identification and quantification of glycoprotein glycosylation. © 2021 Wiley Periodicals LLC. Basic Protocol 1: Characterization of glycans and site occupancy on purified glycoprotein Support Protocol 1: In-gel digestion of glycoproteins Support Protocol 2: Detection of glycoproteins from cells/tissue through glycopeptide enrichment Basic Protocol 2: Acquisition of glycopeptides through high-resolution nano-LC-MS/MS Basic Protocol 3: Identification and quantification of glycopeptides using GlycReSoft.
Keywords: GlycReSoft; bioinformatics; glycomics; glycoproteomics; mass spectrometry.
© 2021 Wiley Periodicals LLC.
Figures

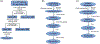




References
-
- Brockhausen I, & Stanley P. (2015). O-GalNAc Glycans. In rd A. Varki, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, & Seeberger PH (Eds.), Essentials of Glycobiology (pp. 113–123). Cold Spring; Harbor (NY).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

