Comparative in silico genome analysis of Clostridium perfringens unravels stable phylogroups with different genome characteristics and pathogenic potential
- PMID: 33762628
- PMCID: PMC7991664
- DOI: 10.1038/s41598-021-86148-8
Comparative in silico genome analysis of Clostridium perfringens unravels stable phylogroups with different genome characteristics and pathogenic potential
Abstract
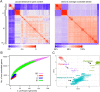
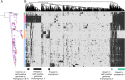
Clostridium perfringens causes a plethora of devastating infections, with toxin production being the underlying mechanism of pathogenicity in various hosts. Genomic analyses of 206 public-available C. perfringens strains´ sequence data identified a substantial degree of genomic variability in respect to episome content, chromosome size and mobile elements. However, the position and order of the local collinear blocks on the chromosome showed a considerable degree of preservation. The strains were divided into five stable phylogroups (I-V). Phylogroup I contained human food poisoning strains with chromosomal enterotoxin (cpe) and a Darmbrand strain characterized by a high frequency of mobile elements, a relatively small genome size and a marked loss of chromosomal genes, including loss of genes encoding virulence traits. These features might correspond to the adaptation of these strains to a particular habitat, causing human foodborne illnesses. This contrasts strains that belong to phylogroup II where the genome size points to the acquisition of genetic material. Most strains of phylogroup II have been isolated from enteric lesions in horses and dogs. Phylogroups III, IV and V are heterogeneous groups containing a variety of different strains, with phylogroup III being the most abundant (65.5%). In conclusion, C. perfringens displays five stable phylogroups reflecting different disease involvements, prompting further studies on the evolution of this highly important pathogen.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

