Protein cleavage influences surface protein presentation in Mycoplasma pneumoniae
- PMID: 33762641
- PMCID: PMC7990945
- DOI: 10.1038/s41598-021-86217-y
Protein cleavage influences surface protein presentation in Mycoplasma pneumoniae
Abstract
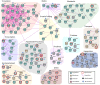
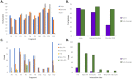
Mycoplasma pneumoniae is a significant cause of pneumonia and post infection sequelae affecting organ sites distant to the respiratory tract are common. It is also a model organism where extensive 'omics' studies have been conducted to gain insight into how minimal genome self-replicating organisms function. An N-terminome study undertaken here identified 4898 unique N-terminal peptides that mapped to 391 (56%) predicted M. pneumoniae proteins. True N-terminal sequences beginning with the initiating methionine (iMet) residue from the predicted Open Reading Frame (ORF) were identified for 163 proteins. Notably, almost half (317; 46%) of the ORFS derived from M. pneumoniae strain M129 are post-translationally modified, presumably by proteolytic processing, because dimethyl labelled neo-N-termini were characterised that mapped beyond the predicted N-terminus. An analysis of the N-terminome describes endoproteolytic processing events predominately targeting tryptic-like sites, though cleavages at negatively charged residues in P1' (D and E) with lysine or serine/alanine in P2' and P3' positions also occurred frequently. Surfaceome studies identified 160 proteins (23% of the proteome) to be exposed on the extracellular surface of M. pneumoniae. The two orthogonal methodologies used to characterise the surfaceome each identified the same 116 proteins, a 72% (116/160) overlap. Apart from lipoproteins, transporters, and adhesins, 93/160 (58%) of the surface proteins lack signal peptides and have well characterised, canonical functions in the cell. Of the 160 surface proteins identified, 134 were also targets of endo-proteolytic processing. These processing events are likely to have profound implications for how the host immune system recognises and responds to M. pneumoniae.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

