NASH limits anti-tumour surveillance in immunotherapy-treated HCC
- PMID: 33762733
- PMCID: PMC8046670
- DOI: 10.1038/s41586-021-03362-0
NASH limits anti-tumour surveillance in immunotherapy-treated HCC
Abstract
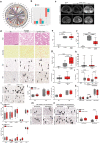
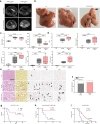
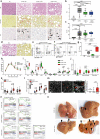
Hepatocellular carcinoma (HCC) can have viral or non-viral causes1-5. Non-alcoholic steatohepatitis (NASH) is an important driver of HCC. Immunotherapy has been approved for treating HCC, but biomarker-based stratification of patients for optimal response to therapy is an unmet need6,7. Here we report the progressive accumulation of exhausted, unconventionally activated CD8+PD1+ T cells in NASH-affected livers. In preclinical models of NASH-induced HCC, therapeutic immunotherapy targeted at programmed death-1 (PD1) expanded activated CD8+PD1+ T cells within tumours but did not lead to tumour regression, which indicates that tumour immune surveillance was impaired. When given prophylactically, anti-PD1 treatment led to an increase in the incidence of NASH-HCC and in the number and size of tumour nodules, which correlated with increased hepatic CD8+PD1+CXCR6+, TOX+, and TNF+ T cells. The increase in HCC triggered by anti-PD1 treatment was prevented by depletion of CD8+ T cells or TNF neutralization, suggesting that CD8+ T cells help to induce NASH-HCC, rather than invigorating or executing immune surveillance. We found similar phenotypic and functional profiles in hepatic CD8+PD1+ T cells from humans with NAFLD or NASH. A meta-analysis of three randomized phase III clinical trials that tested inhibitors of PDL1 (programmed death-ligand 1) or PD1 in more than 1,600 patients with advanced HCC revealed that immune therapy did not improve survival in patients with non-viral HCC. In two additional cohorts, patients with NASH-driven HCC who received anti-PD1 or anti-PDL1 treatment showed reduced overall survival compared to patients with other aetiologies. Collectively, these data show that non-viral HCC, and particularly NASH-HCC, might be less responsive to immunotherapy, probably owing to NASH-related aberrant T cell activation causing tissue damage that leads to impaired immune surveillance. Our data provide a rationale for stratification of patients with HCC according to underlying aetiology in studies of immunotherapy as a primary or adjuvant treatment.
Conflict of interest statement
M.P. is an investigator for Bayer, BMS, Lilly, and Roche; has received speaker honoraria from Bayer, BMS, Eisai, Lilly, and MSD; is a consultant for Bayer, BMS, Eisai, Ipsen, Lilly, MSD, and Roche; and has received travel support from Bayer and BMS. D.P. works currently for Novo Nordisk. M. Szydlowksa works currently for Astra Zeneca. M.K. received honoraria from BMS as consultant and is an investigator for AstraZeneca and BMS. A.V. has served as consultant for Roche, Bayer, Lilly, BMS, Eisai, and Ipsen; has received speaking fees form Roche, Bayer, Lilly, BMS, Eisai, and Ipsen; and is an investigator for Roche, Bayer, Lilly, BMS, Eisai, and Ipsen. F.H. has received travel support from Bayer, Abbvie, and Gilead. M.P.-R. is an advisor/consultant for Astra Zeneca, Bayer, BMS, Eisai, Ipsen, Lilly, and MSD; has served as a speaker for Bayer, Eisai, and Lilly; and is an investigator for Bayer, BMS, Exelixis, and Lilly. F.F. has received travel support from Abbvie and Novartis. O.W. has served as consultant for Amgen, Bayer, BMS, Celgene, Eisai, Merck, Novartis, Roche, Servier, and Shire; has served as a speaker for Abbvie, Bayer, BMS, Celgene, Falk, Ipsen, Novartis, Roche, and Shire; and has received travel support from Abbvie, BMS, Ipsen, Novartis, and Servier. J.T. has served as consultant for Amgen, Bayer, BMS, Eisai, Lilly, Merck Serono, MSD, Ipsen, and Roche; has received travel support from BMS and Ipsen; has received speaking fees from Amgen, Bayer, BMS, Eisai, Lilly, Merck Serono, MSD, Ipsen, and Roche; and is an investigator for Amgen, Bayer, BMS, Eisai, Lilly, Merck Serono, MSD, Ipsen, and Roche. K.S. has served as consultant for Ipsen and Bayer; and conducts studies for Bayer, Roche, Lilly, MSD, and BMS. H.W. has served as speaker for Bayer, Eisai, and Ipsen; has served as a consultant for Bayer, Eisai, Lilly, BMS, Roche, and Ipsen; and conducts studies for Bayer, Roche, Lilly, MSD, and BMS. A. Weberis an advisor for BMS, Wako, Eisai, Roche, and Amgen. J.C.M. has received consulting honoraria from Abbvie, Bayer, BMS, Eisai, Gilead, Incyte, Intercept and MSD for work performed outside the current study. J.M.L. is receiving research support from Bayer HealthCare Pharmaceuticals, Eisai Inc, Bristol-Myers Squibb, Boehringer-Ingelheim and Ipsen, and consulting fees from Eli Lilly, Bayer HealthCare Pharmaceuticals, Bristol-Myers Squibb, Eisai Inc, Celsion Corporation, Exelixis, Merck, Ipsen, Genentech, Roche, Glycotest, Leerink Swann LLC, Fortress Biotech, Nucleix, Can-Fite Biopharma, Sirtex, Mina Alpha Ltd and AstraZeneca. J.M.S. serves as a consultant for Intercept Pharmaceuticals, Genfit, Gilead Sciences, BMS, Madrigal, Novartis, Pfizer, Roche, and Siemens-Healthineers; and has received research funding from Gilead Sciences. D.J.P. has received lecture fees from ViiV Healthcare and Bayer Healthcare; travel expenses from BMS and Bayer Healthcare; consulting fees from Mina Therapeutics, EISAI, Roche and Astra Zeneca; and research funding (to institution) from MSD and BMS. J.-F.D. has served on advisory committees for Abbvie, Bayer, Bristol-Myers Squibb, Falk, Genfit, Genkyotex, Gilead Sciences, HepaRegenix, Intercept, Lilly, Merck, and Novartis; and has spoken or taught at Bayer, Bristol-Myers Squibb, Intercept, Genfit, Gilead Sciences, Novartis, and Roche. L.R. has received consulting fees from Amgen, ArQule, Astra Zeneca, Basilea, Bayer, Celgene, Eisai, Exelixis, Hengrui, Incyte, Ipsen, Lilly, MSD, Nerviano Medical Sciences, Roche, and Sanofi; lectures fees from AbbVie, Amgen, Eisai, Gilead, Incyte, Ipsen, Lilly, Roche, Sanofi; travel expenses from Ipsen; and institutional research funding from Agios, ARMO BioSciences, AstraZeneca, BeiGene, Eisai, Exelixis, Fibrogen, Incyte, Ipsen, Lilly, MSD, and Roche. N.P. has received consulting fees from Amgen, Merck Serono, and Servier; lectures fees from AbbVie, Gilead and Lilly; travel expenses from Amgen and ArQule; and institutional research funding from Basilea, Merck Serono and Servier. T.P. has received institutional research funding from Lilly. D.G.D. has received consultant fees from Bayer, Simcere, Surface Oncology and BMS; and research grants from Bayer, Exelixis and BMS. The remaining authors declare no competing interests.
Figures











Comment in
-
T cell drivers in NASH-HCC.Nat Rev Cancer. 2021 Jun;21(6):341. doi: 10.1038/s41568-021-00362-0. Nat Rev Cancer. 2021. PMID: 33864034 No abstract available.
-
T cells in NASH and liver cancer: pathology and immunotherapy.Nat Rev Gastroenterol Hepatol. 2021 Jun;18(6):367. doi: 10.1038/s41575-021-00461-1. Nat Rev Gastroenterol Hepatol. 2021. PMID: 33948024 No abstract available.
-
The paradox of immunotherapy in NASH-HCC.Signal Transduct Target Ther. 2021 Jun 10;6(1):228. doi: 10.1038/s41392-021-00654-9. Signal Transduct Target Ther. 2021. PMID: 34112767 Free PMC article. No abstract available.
-
Hepatocellular Carcinoma - Origins and Outcomes.N Engl J Med. 2021 Jul 15;385(3):280-282. doi: 10.1056/NEJMcibr2106594. N Engl J Med. 2021. PMID: 34260842 No abstract available.
-
Lack of response to immunotherapy in non-alcoholic steatohepatitis related hepatocellular carcinoma.Hepatobiliary Surg Nutr. 2021 Aug;10(4):522-525. doi: 10.21037/hbsn-21-203. Hepatobiliary Surg Nutr. 2021. PMID: 34430534 Free PMC article. No abstract available.
-
The paradox role of cytotoxic T-lymphocytes in NAFLD-associated hepatocellular carcinoma.Hepatobiliary Surg Nutr. 2021 Oct;10(5):705-707. doi: 10.21037/hbsn-21-243. Hepatobiliary Surg Nutr. 2021. PMID: 34760980 Free PMC article. No abstract available.
-
Fatty liver progression and carcinogenesis: Beware of pathogenic T cells.Med. 2021 May 14;2(5):453-455. doi: 10.1016/j.medj.2021.04.020. Med. 2021. PMID: 35590223
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

