Revealing the mechanism of SARS-CoV-2 spike protein binding with ACE2
- PMID: 33762895
- PMCID: PMC7983027
- DOI: 10.1109/MCSE.2020.3015511
Revealing the mechanism of SARS-CoV-2 spike protein binding with ACE2
Abstract
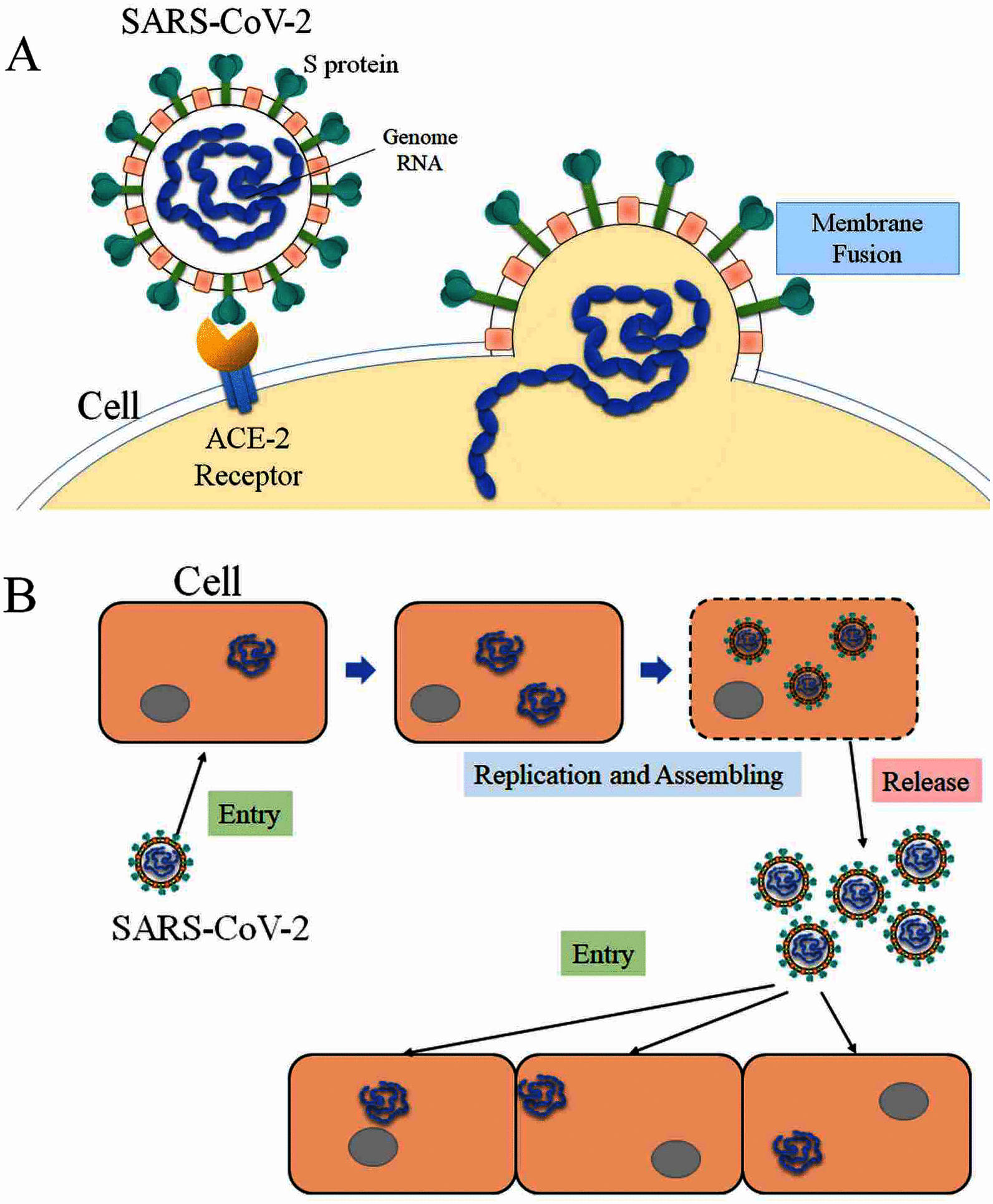
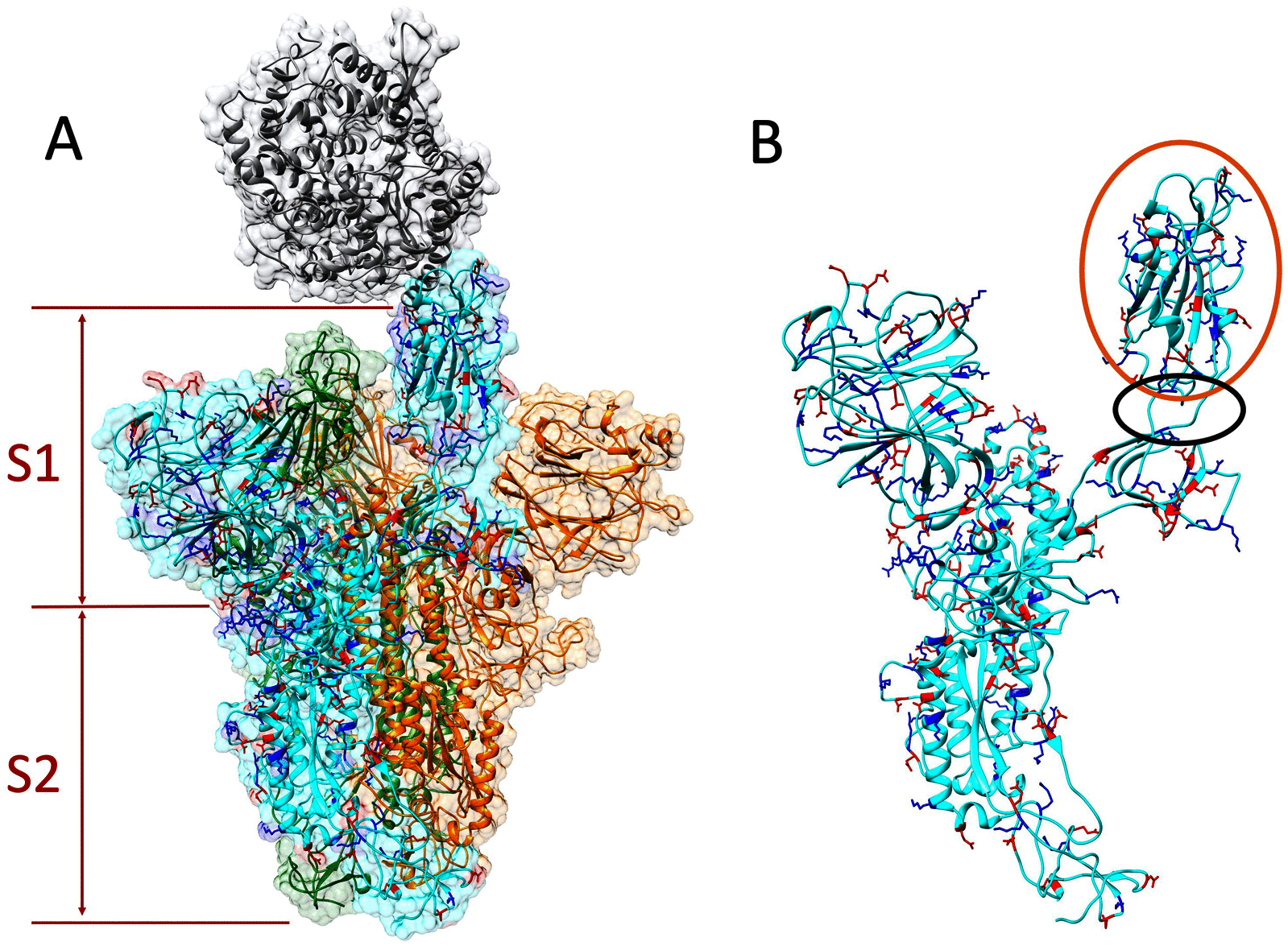
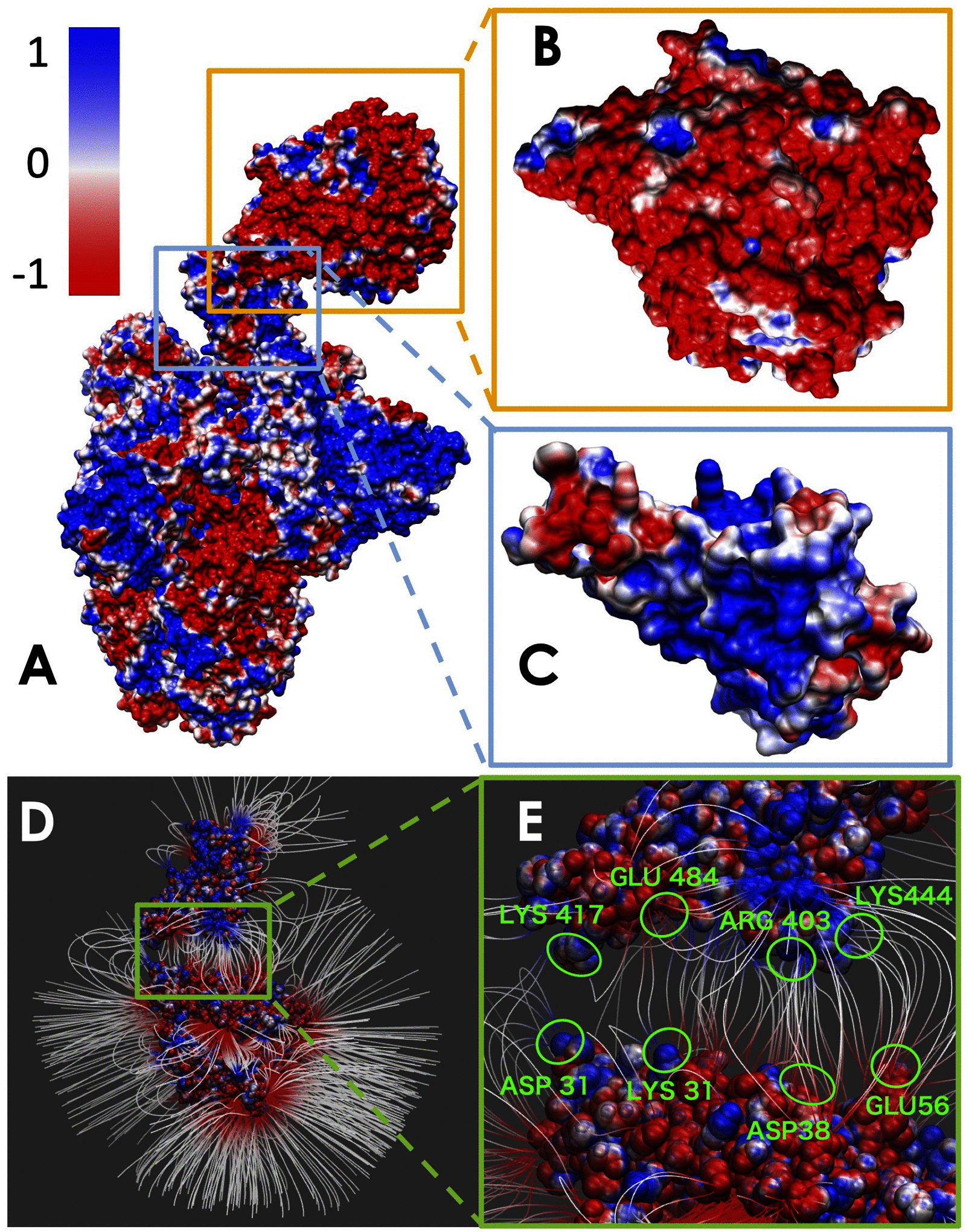
A large population in the world has been infected by COVID-19. Understanding the mechanisms of Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2) is important for management and treatment of the COVID-19. When it comes to the infection process, one of the most important proteins in SARS-CoV-2 is the spike (S) protein, which is able to bind to human Angiotensin-Converting Enzyme 2 (ACE2) and initializes the entry of the host cell. In this study, we implemented multi-scale computational approaches to study the electrostatic features of the interfaces of the SARS-CoV-2 S protein Receptor Binding Domain (RBD) and ACE2. The simulations and analyses were performed on high-performance computing resources in Texas Advanced Computing Center (TACC). Our study identified key residues on the SARS-CoV-2, which can be used as targets for future drug design. The results shed lights on future drug design and therapeutic targets for COVID-19.
Keywords: ACE2; COVID-19; Computational biophysics; SARS-CoV-2; coronavirus; protein-protein interactions; spike protein.
Figures

References
-
- World Health Organization, Coronavirus Disease 2019 (COVID-19): Situation Report, 85, 2020.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
