PI3Kδ Forms Distinct Multiprotein Complexes at the TCR Signalosome in Naïve and Differentiated CD4+ T Cells
- PMID: 33763075
- PMCID: PMC7982423
- DOI: 10.3389/fimmu.2021.631271
PI3Kδ Forms Distinct Multiprotein Complexes at the TCR Signalosome in Naïve and Differentiated CD4+ T Cells
Abstract
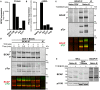
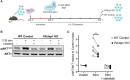
Phosphoinositide 3-kinases (PI3Ks) play a central role in adaptive immunity by transducing signals from the T cell antigen receptor (TCR) via production of PIP3. PI3Kδ is a heterodimer composed of a p110δ catalytic subunit associated with a p85α or p85β regulatory subunit and is preferentially engaged by the TCR upon T cell activation. The molecular mechanisms leading to PI3Kδ recruitment and activation at the TCR signalosome remain unclear. In this study, we have used quantitative mass spectrometry, biochemical approaches and CRISPR-Cas9 gene editing to uncover the p110δ interactome in primary CD4+ T cells. Moreover, we have determined how the PI3Kδ interactome changes upon the differentiation of small naïve T cells into T cell blasts expanded in the presence of IL-2. Our interactomic analyses identified multiple constitutive and inducible PI3Kδ-interacting proteins, some of which were common to naïve and previously-activated T cells. Our data reveals that PI3Kδ rapidly interacts with as many as seven adaptor proteins upon TCR engagement, including the Gab-family proteins, GAB2 and GAB3, a CD5-CBL signalosome and the transmembrane proteins ICOS and TRIM. Our results also suggest that PI3Kδ pre-forms complexes with the adaptors SH3KBP1 and CRKL in resting cells that could facilitate the localization and activation of p110δ at the plasma membrane by forming ternary complexes during early TCR signalling. Furthermore, we identify interactions that were not previously known to occur in CD4+ T cells, involving BCAP, GAB3, IQGAP3 and JAML. We used CRISPR-Cas9-mediated gene knockout in primary T cells to confirm that BCAP is a positive regulator of PI3K-AKT signalling in CD4+ T cell blasts. Overall, our results provide evidence for a large protein network that regulates the recruitment and activation of PI3Kδ in T cells. Finally, this work shows how the PI3Kδ interactome is remodeled as CD4+ T cells differentiate from naïve T cells to activated T cell blasts. These activated T cells upregulate additional PI3Kδ adaptor proteins, including BCAP, GAB2, IQGAP3 and ICOS. This rewiring of TCR-PI3K signalling that occurs upon T cell differentiation may serve to reduce the threshold of activation and diversify the inputs for the PI3K pathway in effector T cells.
Keywords: CD4+ T cells; CRISPR-Cas9; PI3K; TCR signalling; interactomics; p110δ.
Copyright © 2021 Luff, Wojdyla, Oxley, Chessa, Hudson, Hawkins, Stephens, Barry and Okkenhaug.
Conflict of interest statement
STB and KH were employed by the company AstraZeneca. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Rameh LE, Cantley LC. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. (1999) 274:8347–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/B/000C0409/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/R000409/1/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/0000H235/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BBS/E/B/000L0799/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

