LMO7 as an Unrecognized Factor Promoting Pancreatic Cancer Progression and Metastasis
- PMID: 33763427
- PMCID: PMC7982467
- DOI: 10.3389/fcell.2021.647387
LMO7 as an Unrecognized Factor Promoting Pancreatic Cancer Progression and Metastasis
Abstract
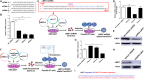
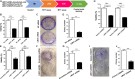
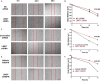
Pancreatic cancer (PC) is one of the most lethal human malignancies without effective treatment. In an effort to discover key genes and molecular pathways underlying PC growth, we have identified LIM domain only 7 (LMO7) as an under-investigated molecule, which highly expresses in primary and metastatic human and mouse PC with the potential of impacting PC tumorigenesis and metastasis. Using genetic methods with siRNA, shRNA, and CRISPR-Cas9, we have successfully generated stable mouse PC cells with LMO7 knockdown or knockout. Using these cells with loss of LMO7 function, we have demonstrated that intrinsic LMO7 defect significantly suppresses PC cell proliferation, anchorage-free colony formation, and mobility in vitro and slows orthotopic PC tumor growth and metastasis in vivo. Mechanistic studies demonstrated that loss of LMO7 function causes PC cell-cycle arrest and apoptosis. These data indicate that LMO7 functions as an independent and unrecognized druggable factor significantly impacting PC growth and metastasis, which could be harnessed for developing a new targeted therapy for PC.
Keywords: CRISPR-Cas9; LIM domain only 7 (LMO7); apoptosis; cell cycle; pancreatic cancer.
Copyright © 2021 Liu, Yuan, Zhou, Wang, Qi, Bernal, Avella, Kaifi, Kimchi, Timothy, Cheng, Miao, Jiang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

