A platform for post-translational spatiotemporal control of cellular proteins
- PMID: 33763602
- PMCID: PMC7976946
- DOI: 10.1093/synbio/ysab002
A platform for post-translational spatiotemporal control of cellular proteins
Abstract
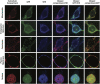
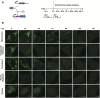
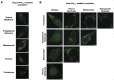
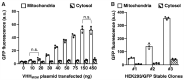
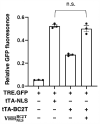
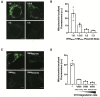
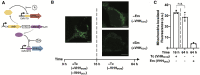
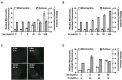
Mammalian cells process information through coordinated spatiotemporal regulation of proteins. Engineering cellular networks thus relies on efficient tools for regulating protein levels in specific subcellular compartments. To address the need to manipulate the extent and dynamics of protein localization, we developed a platform technology for the target-specific control of protein destination. This platform is based on bifunctional molecules comprising a target-specific nanobody and universal sequences determining target subcellular localization or degradation rate. We demonstrate that nanobody-mediated localization depends on the expression level of the target and the nanobody, and the extent of target subcellular localization can be regulated by combining multiple target-specific nanobodies with distinct localization or degradation sequences. We also show that this platform for nanobody-mediated target localization and degradation can be regulated transcriptionally and integrated within orthogonal genetic circuits to achieve the desired temporal control over spatial regulation of target proteins. The platform reported in this study provides an innovative tool to control protein subcellular localization, which will be useful to investigate protein function and regulate large synthetic gene circuits.
Keywords: degradation; localization; mammalian genetic circuits; nanobody; orthogonal protein regulation.
© The Author(s) 2021. Published by Oxford University Press.
Figures
Similar articles
-
Design of Oscillatory Networks through Post-Translational Control of Network Components.Synth Biol Eng. 2023 Jun;1(1):10004. doi: 10.35534/sbe.2023.10004. Epub 2023 Mar 13. Synth Biol Eng. 2023. PMID: 38590452 Free PMC article.
-
Input-dependent post-translational control of the reporter output enhances dynamic resolution of mammalian signaling systems.Methods Enzymol. 2019;622:1-27. doi: 10.1016/bs.mie.2019.02.013. Epub 2019 Mar 12. Methods Enzymol. 2019. PMID: 31155049
-
Engineering and characterization of GFP-targeting nanobody: Expression, purification, and post-translational modification analysis.Protein Expr Purif. 2024 Sep;221:106501. doi: 10.1016/j.pep.2024.106501. Epub 2024 May 21. Protein Expr Purif. 2024. PMID: 38782081
-
Nanobody-Based Probes for Subcellular Protein Identification and Visualization.Front Cell Neurosci. 2020 Nov 2;14:573278. doi: 10.3389/fncel.2020.573278. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33240044 Free PMC article. Review.
-
Recombinant expression of nanobodies and nanobody-derived immunoreagents.Protein Expr Purif. 2020 Aug;172:105645. doi: 10.1016/j.pep.2020.105645. Epub 2020 Apr 11. Protein Expr Purif. 2020. PMID: 32289357 Free PMC article. Review.
Cited by
-
A programmable reaction-diffusion system for spatiotemporal cell signaling circuit design.Cell. 2024 Jan 18;187(2):345-359.e16. doi: 10.1016/j.cell.2023.12.007. Epub 2024 Jan 4. Cell. 2024. PMID: 38181787 Free PMC article.
-
Targeted protein relocalization via protein transport coupling.Nature. 2024 Sep;633(8031):941-951. doi: 10.1038/s41586-024-07950-8. Epub 2024 Sep 18. Nature. 2024. PMID: 39294374 Free PMC article.
References
-
- Berg J., Tymoczko J., Stryer L. (2002) Biochemistry, 5th edn. W. H. Freeman and Company, New York.
-
- Bauer N.C., Doetsch P.W., Corbett A.H. (2015) Mechanisms regulating protein localization. Traffic, 16, 1039–1061. - PubMed
-
- Weake V.M., Workman J.L. (2010) Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet., 11, 426–437. [10.1038/nrg2781]. - PubMed
-
- Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. (2018) The human transcription factors. Cell, 172, 650–665. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
