Genomic Context Differs Between Human Dilated Cardiomyopathy and Hypertrophic Cardiomyopathy
- PMID: 33764162
- PMCID: PMC8174318
- DOI: 10.1161/JAHA.120.019944
Genomic Context Differs Between Human Dilated Cardiomyopathy and Hypertrophic Cardiomyopathy
Abstract
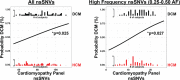
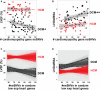
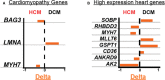
Background Inherited cardiomyopathies display variable penetrance and expression, and a component of phenotypic variation is genetically determined. To evaluate the genetic contribution to this variable expression, we compared protein coding variation in the genomes of those with hypertrophic cardiomyopathy (HCM) and dilated cardiomyopathy (DCM). Methods and Results Nonsynonymous single-nucleotide variants (nsSNVs) were ascertained using whole genome sequencing from familial cases of HCM (n=56) or DCM (n=70) and correlated with echocardiographic information. Focusing on nsSNVs in 102 genes linked to inherited cardiomyopathies, we correlated the number of nsSNVs per person with left ventricular measurements. Principal component analysis and generalized linear models were applied to identify the probability of cardiomyopathy type as it related to the number of nsSNVs in cardiomyopathy genes. The probability of having DCM significantly increased as the number of cardiomyopathy gene nsSNVs per person increased. The increase in nsSNVs in cardiomyopathy genes significantly associated with reduced left ventricular ejection fraction and increased left ventricular diameter for individuals carrying a DCM diagnosis, but not for those with HCM. Resampling was used to identify genes with aberrant cumulative allele frequencies, identifying potential modifier genes for cardiomyopathy. Conclusions Participants with DCM had more nsSNVs per person in cardiomyopathy genes than participants with HCM. The nsSNV burden in cardiomyopathy genes did not correlate with the probability or manifestation of left ventricular measures in HCM. These findings support the concept that increased variation in cardiomyopathy genes creates a genetic background that predisposes to DCM and increased disease severity.
Keywords: dilated cardiomyopathy; hypertrophic cardiomyopathy; modifier genes; variable expressivity; variant burden.
Conflict of interest statement
Dr McNally serves as a consultant to Invitae and Tenaya Therapeutics. The remaining authors have no disclosures to report.
Figures

References
-
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, et al. Heart disease and stroke statistics–2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–e322. DOI: 10.1161/CIR.0000000000000152 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

