Cytoplasmic HYL1 modulates miRNA-mediated translational repression
- PMID: 33764452
- PMCID: PMC8290291
- DOI: 10.1093/plcell/koab090
Cytoplasmic HYL1 modulates miRNA-mediated translational repression
Abstract
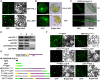
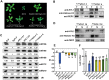
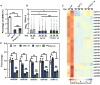
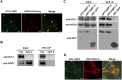
MicroRNAs (miRNAs) control various biological processes by repressing target mRNAs. In plants, miRNAs mediate target gene repression via both mRNA cleavage and translational repression. However, the mechanism underlying this translational repression is poorly understood. Here, we found that Arabidopsis thaliana HYPONASTIC LEAVES1 (HYL1), a core component of the miRNA processing machinery, regulates miRNA-mediated mRNA translation but not miRNA biogenesis when it localized in the cytoplasm. Cytoplasmic HYL1 localizes to the endoplasmic reticulum and associates with ARGONAUTE1 (AGO1) and ALTERED MERISTEM PROGRAM1. In the cytoplasm, HYL1 monitors the distribution of AGO1 onto polysomes, binds to the mRNAs of target genes, represses their translation, and partially rescues the phenotype of the hyl1 null mutant. This study uncovered another function of HYL1 and provides insight into the mechanism of plant gene regulation.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



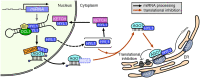
Similar articles
-
The Intrinsically Disordered Protein CARP9 Bridges HYL1 to AGO1 in the Nucleus to Promote MicroRNA Activity.Plant Physiol. 2020 Sep;184(1):316-329. doi: 10.1104/pp.20.00258. Epub 2020 Jul 7. Plant Physiol. 2020. PMID: 32636339 Free PMC article.
-
KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis.Proc Natl Acad Sci U S A. 2017 Apr 11;114(15):4011-4016. doi: 10.1073/pnas.1619755114. Epub 2017 Mar 27. Proc Natl Acad Sci U S A. 2017. PMID: 28348234 Free PMC article.
-
MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis.Cell. 2013 Apr 25;153(3):562-74. doi: 10.1016/j.cell.2013.04.005. Cell. 2013. PMID: 23622241 Free PMC article.
-
Post-Translational Regulation of miRNA Pathway Components, AGO1 and HYL1, in Plants.Mol Cells. 2016 Aug 31;39(8):581-6. doi: 10.14348/molcells.2016.0085. Epub 2016 Jul 20. Mol Cells. 2016. PMID: 27440184 Free PMC article. Review.
-
Argonautes compete for miR165/166 to regulate shoot apical meristem development.Curr Opin Plant Biol. 2012 Dec;15(6):652-8. doi: 10.1016/j.pbi.2012.05.007. Epub 2012 Jun 21. Curr Opin Plant Biol. 2012. PMID: 22727764 Free PMC article. Review.
Cited by
-
Function and regulation of plant ARGONAUTE proteins in response to environmental challenges: a review.PeerJ. 2024 Mar 26;12:e17115. doi: 10.7717/peerj.17115. eCollection 2024. PeerJ. 2024. PMID: 38560454 Free PMC article. Review.
-
Switching action modes of miR408-5p mediates auxin signaling in rice.Nat Commun. 2024 Mar 21;15(1):2525. doi: 10.1038/s41467-024-46765-z. Nat Commun. 2024. PMID: 38514635 Free PMC article.
-
Unlocking the molecular basis of wheat straw composition and morphological traits through multi-locus GWAS.BMC Plant Biol. 2022 Nov 8;22(1):519. doi: 10.1186/s12870-022-03900-6. BMC Plant Biol. 2022. PMID: 36344939 Free PMC article.
-
Heat stress triggers enhanced nuclear localization of HYPONASTIC LEAVES 1 to regulate microRNA biogenesis and thermotolerance in plants.Plant Cell. 2025 Jun 4;37(6):koaf092. doi: 10.1093/plcell/koaf092. Plant Cell. 2025. PMID: 40266261
-
MicroRNA398: A Master Regulator of Plant Development and Stress Responses.Int J Mol Sci. 2022 Sep 16;23(18):10803. doi: 10.3390/ijms231810803. Int J Mol Sci. 2022. PMID: 36142715 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

