Dissemination and evolution of SARS-CoV-2 in the early pandemic phase in South America
- PMID: 33764553
- PMCID: PMC8250755
- DOI: 10.1002/jmv.26967
Dissemination and evolution of SARS-CoV-2 in the early pandemic phase in South America
Abstract
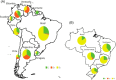
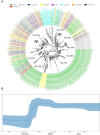
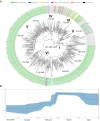
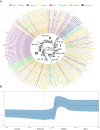
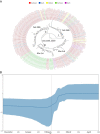
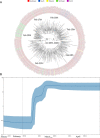
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic spread rapidly and this scenario is concerning in South America, mainly in Brazil with more than seven million cases of infection. Three major pandemic lineages/clades could be identified along with SARS-CoV-2 dissemination (G, GR, and GH) in the Americas. These clades differ according to their genomic characteristics, virulence, and spreading times. The present study describes the main clades and the respective temporal spreading analyses based on SARS-CoV-2 whole-genome sequences (WGS) from South America, obtained in the early pandemic phase (from March 1 to May 31 in 2020). SARS-CoV-2 WGSs with available information from country and year of sampling were obtained from different countries and the main clades were identified and analyzed independently with a Bayesian approach. The results demonstrated the prevalence of clades GR (n = 842; 54.6%), G (n = 529; 34.3%), and GH (n = 171; 11.1%). The frequencies of the clades were significantly different between South American countries. Clade G was the most prevalent in Ecuador, Suriname, and Uruguay, clade GR in Argentina, Brazil, and Peru, and clade GH in Colombia. The phylodynamic analysis indicated that all these main lineages increased viral spreading from February to early March and after an evolutionary stationary phase was observed. The decrease observed in the virus dissemination was directly associated to the reduction of social movement after March. In conclusion, these data demonstrated the current predominance of clades G, GR, and GH in South America because of the early dissemination of them in the first pandemic phase in South America.
Keywords: SARS coronavirus; dissemination; pandemic.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that htere are no conflicts of interests.
Figures
References
-
- GISAID Initiative. COVID‐19 dashboard by the Center for Systems Science and Engineering (CSSE) at John Hopkins University. 2020. https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594.... Accessed November 12, 2020.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

