Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation
- PMID: 33764843
- PMCID: PMC8726636
- DOI: 10.1080/15548627.2021.1901204
Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation
Abstract
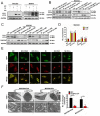
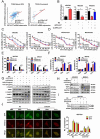
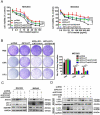
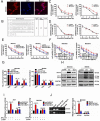
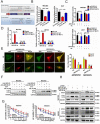
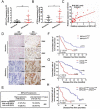
Chemotherapy is currently the main treatment for unresectable or advanced postoperative gastric cancers. However, its efficacy is negatively affected by the occurrence of chemoresistance, which severely affects patient prognosis. Recently, dysregulation in autophagy has been suggested as a potential mechanism for chemoresistence, and long noncoding RNA (lncRNA) also shows its regulatory role in cancer drug resistance. Using RNA sequencing, we found that lncRNA EIF3J-DT was highly expressed in drug-resistant gastric cancer cells. In-vitro and in-vivo experiments showed that EIF3J-DT activated autophagy and induced drug resistance in gastric cancer cells by targeting ATG14. Bioinformatics and experimental results showed that EIF3J-DT regulated the expression of ATG14 through direct binding to enhance stabilization of ATG14 mRNA and via blocking the degradation of ATG14 mRNA through competitively binding with microRNA (miRNA) MIR188-3p. Therefore, EIF3J-DT increased the expression of ATG14, contributing to activation of autophagy and chemoresistance. Furthermore, it was confirmed that EIF3J-DT and ATG14 were highly expressed in gastric cancer patients resistant to chemotherapy, and this was closely associated with patient prognosis. In conclusion, EIF3J-DT is involved in the regulation of autophagy and chemoresistance in gastric cancer cells by targeting ATG14. It may be a suitable new target for enhancing chemosensitivity and improving prognosis.Abbreviations: 3-MA: 3-methyladenine; 5-Fu: 5-fluorouracil; ATG: autophagy related; C-CASP3: cleaved caspase 3; C-CASP7: cleaved caspase 7; C-PARP: cleaved PARP; CQ: chloroquine; CR: complete response; DIG: digoxigenin; ESR1: estrogen receptor 1; FBS: fetal bovine serum; FISH: fluorescence in situ hybridization; IHC: immunohistochemistry; ISH: in situ hybridization; lncRNA: long noncoding RNA; miRNA: microRNA; MUT: mutant; NC: negative control; OXA: oxaliplatin; PBS: phosphate-buffered saline; PD: progressive disease; PFA: paraformaldehyde; PR: partial response; qPCR: quantitative polymerase chain reaction; RAPA: rapamycin; SD: stable disease; TEM: transmission electron microscopy; WT: wild type.
Keywords: ATG14; EIF3J-DT; gastric cancer; autophagy; chemotherapy resistance.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
