Genome-Wide Analysis of Translation in Replicatively Aged Yeast
- PMID: 33765274
- PMCID: PMC8565997
- DOI: 10.1007/978-1-0716-1150-0_6
Genome-Wide Analysis of Translation in Replicatively Aged Yeast
Abstract
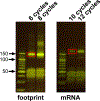
Protein synthesis is an essential process that affects major cellular functions including growth, energy production, cell signaling, and enzymatic reactions. However, how it is impacted by aging and how the translation of specific proteins is changed during the aging process remain understudied. Although yeast is a widely used model for studying eukaryotic aging, analysis of age-related translational changes using ribosome profiling in this organism has been challenging due to the need for isolating large quantities of old cells. Here, we provide a detailed protocol for genome-wide analysis of protein synthesis using ribosome profiling in replicatively aged yeast. By combining genetic enrichment of old cells with the biotin affinity purification step, this method allows large-scale isolation of aged cells sufficient for generating ribosome profiling libraries. We also describe a strategy for normalization of samples using a spike-in with worm lysates that permits quantitative comparison of absolute translation levels between young and old cells.
Keywords: Aging; Mother enrichment program; Protein translation; Ribo-Seq; Saccharomyces cerevisiae; Spike-in; Yeast.
Figures

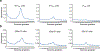
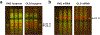
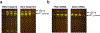
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

