Vaccinomic approach for novel multi epitopes vaccine against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)
- PMID: 33765919
- PMCID: PMC7992937
- DOI: 10.1186/s12865-021-00412-0
Vaccinomic approach for novel multi epitopes vaccine against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)
Abstract
Background: The spread of a novel coronavirus termed severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in China and other countries is of great concern worldwide with no effective vaccine. This study aimed to design a novel vaccine construct against SARS-CoV-2 from the spike S protein and orf1ab polyprotein using immunoinformatics tools. The vaccine was designed from conserved epitopes interacted against B and T lymphocytes by the combination of highly immunogenic epitopes with suitable adjuvant and linkers.
Results: The proposed vaccine composed of 526 amino acids and was shown to be antigenic in Vaxigen server (0.6194) and nonallergenic in Allertop server. The physiochemical properties of the vaccine showed isoelectric point of 10.19. The instability index (II) was 31.25 classifying the vaccine as stable. Aliphatic index was 84.39 and the grand average of hydropathicity (GRAVY) was - 0.049 classifying the vaccine as hydrophilic. Vaccine tertiary structure was predicted, refined and validated to assess the stability of the vaccine via Ramachandran plot and ProSA-web servers. Moreover, solubility of the vaccine construct was greater than the average solubility provided by protein sol and SOLpro servers indicating the solubility of the vaccine construct. Disulfide engineering was performed to reduce the high mobile regions in the vaccine to enhance stability. Docking of the vaccine construct with TLR4 demonstrated efficient binding energy with attractive binding energy of - 338.68 kcal/mol and - 346.89 kcal/mol for TLR4 chain A and chain B respectively. Immune simulation significantly provided high levels of immunoglobulins, T-helper cells, T-cytotoxic cells and INF-γ. Upon cloning, the vaccine protein was reverse transcribed into DNA sequence and cloned into pET28a(+) vector to ensure translational potency and microbial expression.
Conclusion: A unique vaccine construct from spike S protein and orf1ab polyprotein against B and T lymphocytes was generated with potential protection against the pandemic. The present study might assist in developing a suitable therapeutics protocol to combat SARSCoV-2 infection.
Keywords: B-lymphocytes; Multiepitopes vaccine; SARS CoV-2; Spike S protein; T-lymphocytes; orf1ab polyprotein.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


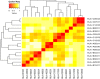
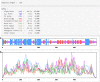
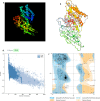
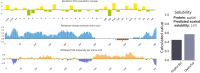


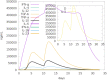
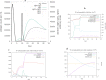

References
-
- World Health Organization. WHO director-general’s remarks at the media briefing on 2019-nCoV on 11 February 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at.... Accessed 13 Feb 2020.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

