Metagenomic insights into the taxonomy, function, and dysbiosis of prokaryotic communities in octocorals
- PMID: 33766108
- PMCID: PMC7993494
- DOI: 10.1186/s40168-021-01031-y
Metagenomic insights into the taxonomy, function, and dysbiosis of prokaryotic communities in octocorals
Abstract
Background: In octocorals (Cnidaria Octocorallia), the functional relationship between host health and its symbiotic consortium has yet to be determined. Here, we employed comparative metagenomics to uncover the distinct functional and phylogenetic features of the microbiomes of healthy Eunicella gazella, Eunicella verrucosa, and Leptogorgia sarmentosa tissues, in contrast with the microbiomes found in seawater and sediments. We further explored how the octocoral microbiome shifts to a pathobiome state in E. gazella.
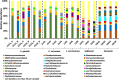
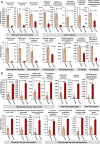
Results: Multivariate analyses based on 16S rRNA genes, Clusters of Orthologous Groups of proteins (COGs), Protein families (Pfams), and secondary metabolite-biosynthetic gene clusters annotated from 20 Illumina-sequenced metagenomes each revealed separate clustering of the prokaryotic communities of healthy tissue samples of the three octocoral species from those of necrotic E. gazella tissue and surrounding environments. While the healthy octocoral microbiome was distinguished by so-far uncultivated Endozoicomonadaceae, Oceanospirillales, and Alteromonadales phylotypes in all host species, a pronounced increase of Flavobacteriaceae and Alphaproteobacteria, originating from seawater, was observed in necrotic E. gazella tissue. Increased abundances of eukaryotic-like proteins, exonucleases, restriction endonucleases, CRISPR/Cas proteins, and genes encoding for heat-shock proteins, inorganic ion transport, and iron storage distinguished the prokaryotic communities of healthy octocoral tissue regardless of the host species. An increase of arginase and nitric oxide reductase genes, observed in necrotic E. gazella tissues, suggests the existence of a mechanism for suppression of nitrite oxide production by which octocoral pathogens may overcome the host's immune system.
Conclusions: This is the first study to employ primer-less, shotgun metagenome sequencing to unveil the taxonomic, functional, and secondary metabolism features of prokaryotic communities in octocorals. Our analyses reveal that the octocoral microbiome is distinct from those of the environmental surroundings, is host genus (but not species) specific, and undergoes large, complex structural changes in the transition to the dysbiotic state. Host-symbiont recognition, abiotic-stress response, micronutrient acquisition, and an antiviral defense arsenal comprising multiple restriction endonucleases, CRISPR/Cas systems, and phage lysogenization regulators are signatures of prokaryotic communities in octocorals. We argue that these features collectively contribute to the stabilization of symbiosis in the octocoral holobiont and constitute beneficial traits that can guide future studies on coral reef conservation and microbiome therapy. Video Abstract.
Keywords: Eunicella; Gorgonians; Holobiont; Host-microbe interactions; Leptogorgia; Necrosis; Secondary metabolism; Symbiosis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Margulis L. Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. Cambridge: MIT Press; 1991. - PubMed
-
- Jeon Y, Park SG, Lee N, Weber JA, Kim H-S, Hwang S-J, Woo S, Kim H-M, Bhak Y, Jeon S, Lee N, Jo Y, Blazyte A, Ryu T, Cho YS, Kim H, Lee JH, Yim HS, Bhak J, Yum S. The draft genome of an octocoral, Dendronephthya gigantea. Genome Biol Evol. 2019;11(3):949–953. doi: 10.1093/gbe/evz043. - DOI - PMC - PubMed
-
- Williams GC, Cairns SD. Systematic list of valid octocoral genera. 2019.
-
- Pérez CD, de Moura NB, Cordeiro RT, Williams GC. Diversity and distribution of Octocorallia. In: Goffredo S, Dubinsky Z, editors. The Cnidaria, past, present and future: the world of Medusa and her sisters. Springer International Publishing; 2016. pp. 109–123.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

