Calmodulin influences MAPK signaling by binding KSR1
- PMID: 33766558
- PMCID: PMC8079274
- DOI: 10.1016/j.jbc.2021.100577
Calmodulin influences MAPK signaling by binding KSR1
Abstract
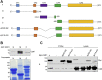
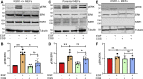
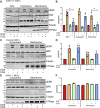
The mitogen-activated protein kinase (MAPK) cascade is a fundamental signaling pathway that regulates cell fate decisions in response to external stimuli. Several scaffold proteins bind directly to kinase components of this pathway and regulate their activation by growth factors. One of the best studied MAPK scaffolds is kinase suppressor of Ras1 (KSR1), which is induced by epidermal growth factor (EGF) to translocate to the plasma membrane where it activates extracellular signal-regulated kinase (ERK). While Ca2+ has been shown to modulate MAPK signaling, the molecular mechanisms by which this occurs are incompletely understood. Here we tested the hypothesis that Ca2+ alters MAPK activity at least in part via KSR1. Using several approaches, including fusion proteins, immunoprecipitation, confocal microscopy, and a cell-permeable chemical inhibitor, we investigated the functional interaction between KSR1 and calmodulin. In vitro analysis with pure proteins reveals that calmodulin binds directly to KSR1. Moreover, endogenous calmodulin and KSR1 co-immunoprecipitate from mammalian cell lysates. Importantly, Ca2+ is required for the association between calmodulin and KSR1, both in vitro and in cells. The cell-permeable calmodulin antagonist CGS9343B significantly reduced activation of ERK by EGF in mouse embryo fibroblasts that overexpress KSR1, but not in control cells. Moreover, CGS9343B impaired the ability of EGF to induce KSR1 translocation to the plasma membrane and to stimulate formation of KSR1-ERK and KSR1-pERK (phosphorylated ERK) complexes in cells. Collectively, our data identify a previously unrecognized mechanism by which the scaffold protein KSR1 couples Ca2+ and calmodulin signaling to the MAPK cascade.
Keywords: calcium; calmodulin; kinase suppressor of Ras1 (KSR1); mitogen-activated protein kinase (MAPK); protein–protein interaction; signaling.
Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
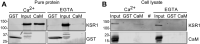



References
-
- Lavoie H., Gagnon J., Therrien M. ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 2020;21:607–632. - PubMed
-
- Morrison D.K., Davis R.J. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. - PubMed
-
- Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 2005;6:827–837. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

