Artificial intelligence in OCT angiography
- PMID: 33766775
- PMCID: PMC8455727
- DOI: 10.1016/j.preteyeres.2021.100965
Artificial intelligence in OCT angiography
Abstract
Optical coherence tomographic angiography (OCTA) is a non-invasive imaging modality that provides three-dimensional, information-rich vascular images. With numerous studies demonstrating unique capabilities in biomarker quantification, diagnosis, and monitoring, OCTA technology has seen rapid adoption in research and clinical settings. The value of OCTA imaging is significantly enhanced by image analysis tools that provide rapid and accurate quantification of vascular features and pathology. Today, the most powerful image analysis methods are based on artificial intelligence (AI). While AI encompasses a large variety of techniques, machine-learning-based, and especially deep-learning-based, image analysis provides accurate measurements in a variety of contexts, including different diseases and regions of the eye. Here, we discuss the principles of both OCTA and AI that make their combination capable of answering new questions. We also review contemporary applications of AI in OCTA, which include accurate detection of pathologies such as choroidal neovascularization, precise quantification of retinal perfusion, and reliable disease diagnosis.
Keywords: Artificial intelligence; Deep learning; Image analysis; OCT Angiography.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
All relevant disclosures are given below, and will be included in a future manuscript version that includes author details.
Disclosures
Oregon Health & Science University (OHSU) and Drs. Jia and Huang have a financial interest in Optovue Inc. These potential conflicts of interest have been reviewed and are managed by OHSU. The other authors do not have any potential financial conflicts of interest.
Figures


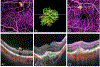
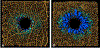
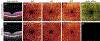
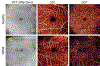








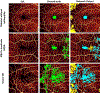
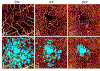
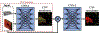

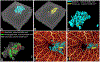

References
-
- Agemy SA, Scripsema NK, Shah CM, Chui T, Garcia PM, Lee JG, Gentile RC, Hsiao YS, Zhou Q, Ko T, Rosen RB, 2015. Retinal vascular perfusion density mapping using optical coherence tomography angiography in normals and diabetic retinopathy patients. Retina 35, 2353–2363. 10.1097/IAE.0000000000000862 - DOI - PubMed
-
- Ahlers C, Simader C, Geitzenauer W, Stock G, Stetson P, Dastmalchi S, Schmidt-Erfurth U, 2008. Automatic segmentation in three-dimensional analysis of fibrovascular pigmentepithelial detachment using high-definition optical coherence tomography. Br. J. Ophthalmol. 92, 197–203. 10.1136/bjo.2007.120956 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

