Cytomegaloviral determinants of CD8+ T cell programming and RhCMV/SIV vaccine efficacy
- PMID: 33766849
- PMCID: PMC8244349
- DOI: 10.1126/sciimmunol.abg5413
Cytomegaloviral determinants of CD8+ T cell programming and RhCMV/SIV vaccine efficacy
Abstract
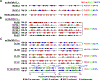
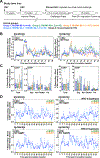
Simian immunodeficiency virus (SIV) insert-expressing, 68-1 rhesus cytomegalovirus (RhCMV/SIV) vectors elicit major histocompatibility complex E (MHC-E)- and MHC-II-restricted, SIV-specific CD8+ T cell responses, but the basis of these unconventional responses and their contribution to demonstrated vaccine efficacy against SIV challenge in the rhesus monkeys (RMs) have not been characterized. We show that these unconventional responses resulted from a chance genetic rearrangement in 68-1 RhCMV that abrogated the function of eight distinct immunomodulatory gene products encoded in two RhCMV genomic regions (Rh157.5/Rh157.4 and Rh158-161), revealing three patterns of unconventional response inhibition. Differential repair of these genes with either RhCMV-derived or orthologous human CMV (HCMV)-derived sequences (UL128/UL130; UL146/UL147) leads to either of two distinct CD8+ T cell response types-MHC-Ia-restricted only or a mix of MHC-II- and MHC-Ia-restricted CD8+ T cells. Response magnitude and functional differentiation are similar to RhCMV 68-1, but neither alternative response type mediated protection against SIV challenge. These findings implicate MHC-E-restricted CD8+ T cell responses as mediators of anti-SIV efficacy and indicate that translation of RhCMV/SIV vector efficacy to humans will likely require deletion of all genes that inhibit these responses from the HCMV/HIV vector.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures






Comment in
-
CMV, MHC-E, and the quest for an unconventional AIDS vaccine.Sci Immunol. 2021 May 14;6(59):eabi5830. doi: 10.1126/sciimmunol.abi5830. Sci Immunol. 2021. PMID: 33990380
References
-
- Jarvis MA, Hansen SG, Nelson JA, Picker LJ, Früh K, in Cytomegaloviruses: From Molecular Pathogenesis to Intervention Reddehase MJ, Ed. (Caister Academic Press, 2013), vol. 2, chap. 21.
-
- Vieira Braga FA, Hertoghs KM, van Lier RA, van Gisbergen KP, Molecular characterization of HCMV-specific immune responses: Parallels between CD8(+) T cells, CD4(+) T cells, and NK cells. Eur J Immunol 45, 2433–2445 (2015). - PubMed
-
- Klenerman P, Oxenius A, T cell responses to cytomegalovirus. Nat Rev Immunol 16, 367–377 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

