The microbiome and human cancer
- PMID: 33766858
- PMCID: PMC8767999
- DOI: 10.1126/science.abc4552
The microbiome and human cancer
Erratum in
-
Erratum for the Review "The microbiome and human cancer" by G. D. Sepich-Poore et al.Science. 2024 Sep 27;385(6716):eadt2260. doi: 10.1126/science.adt2260. Epub 2024 Sep 26. Science. 2024. PMID: 39325913 No abstract available.
Abstract
Microbial roles in cancer formation, diagnosis, prognosis, and treatment have been disputed for centuries. Recent studies have provocatively claimed that bacteria, viruses, and/or fungi are pervasive among cancers, key actors in cancer immunotherapy, and engineerable to treat metastases. Despite these findings, the number of microbes known to directly cause carcinogenesis remains small. Critically evaluating and building frameworks for such evidence in light of modern cancer biology is an important task. In this Review, we delineate between causal and complicit roles of microbes in cancer and trace common themes of their influence through the host's immune system, herein defined as the immuno-oncology-microbiome axis. We further review evidence for intratumoral microbes and approaches that manipulate the host's gut or tumor microbiome while projecting the next phase of experimental discovery.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
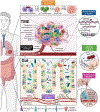

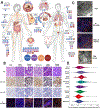
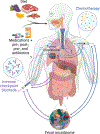
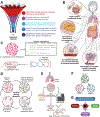
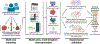
Comment in
-
Roles of the human microbiome in cancer.Hepatobiliary Surg Nutr. 2021 Aug;10(4):558-560. doi: 10.21037/hbsn-21-241. Hepatobiliary Surg Nutr. 2021. PMID: 34430543 Free PMC article. No abstract available.
References
-
- Ebbell B, The Papyrus Ebers: the greatest Egyptian medical document (Levin & Munksgaard, 1937).
-
- Busch W, Aus der Sitzung der medicinischen Section vom 13 November 1867. Berl Klin Wochenschr. 5, 137 (1868).
-
- Fehleisen F, Ueber die Züchtung der Erysipelkokken auf künstlichem Nährboden und ihre Übertragbarkeit auf den Menschen. Dtsch. Med. Wochenschr 8, 553–554 (1882).
-
- Starnes CO, Coley’s toxins in perspective. Nature. 357, 11–12 (1992). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

