A RIPK1-regulated inflammatory microglial state in amyotrophic lateral sclerosis
- PMID: 33766915
- PMCID: PMC8020785
- DOI: 10.1073/pnas.2025102118
A RIPK1-regulated inflammatory microglial state in amyotrophic lateral sclerosis
Abstract
Microglial-derived inflammation has been linked to a broad range of neurodegenerative and neuropsychiatric conditions, including amyotrophic lateral sclerosis (ALS). Using single-cell RNA sequencing, a class of Disease-Associated Microglia (DAMs) have been characterized in neurodegeneration. However, the DAM phenotype alone is insufficient to explain the functional complexity of microglia, particularly with regard to regulating inflammation that is a hallmark of many neurodegenerative diseases. Here, we identify a subclass of microglia in mouse models of ALS which we term RIPK1-Regulated Inflammatory Microglia (RRIMs). RRIMs show significant up-regulation of classical proinflammatory pathways, including increased levels of Tnf and Il1b RNA and protein. We find that RRIMs are highly regulated by TNFα signaling and that the prevalence of these microglia can be suppressed by inhibiting receptor-interacting protein kinase 1 (RIPK1) activity downstream of the TNF receptor 1. These findings help to elucidate a mechanism by which RIPK1 kinase inhibition has been shown to provide therapeutic benefit in mouse models of ALS and may provide an additional biomarker for analysis in ongoing phase 2 clinical trials of RIPK1 inhibitors in ALS.
Keywords: ALS; RIPK1; microglia; neuroinflammation; scRNAseq.
Conflict of interest statement
Competing interest statement: J.Y. is a consultant for Denali Therapeutics and Sanofi. J.Y. and J.M.H. are co-authors on a 2018 Nomenclature review article.
Figures

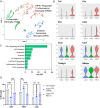
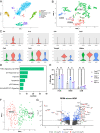
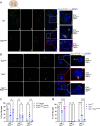
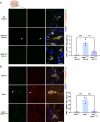
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

