WWTR1(TAZ)- CAMTA1 gene fusion is sufficient to dysregulate YAP/TAZ signaling and drive epithelioid hemangioendothelioma tumorigenesis
- PMID: 33766982
- PMCID: PMC8015722
- DOI: 10.1101/gad.348220.120
WWTR1(TAZ)- CAMTA1 gene fusion is sufficient to dysregulate YAP/TAZ signaling and drive epithelioid hemangioendothelioma tumorigenesis
Abstract
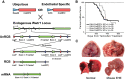
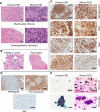
Epithelioid hemangioendothelioma (EHE) is a genetically homogenous vascular sarcoma that is a paradigm for TAZ dysregulation in cancer. EHE harbors a WWTR1(TAZ)-CAMTA1 gene fusion in >90% of cases, 45% of which have no other genetic alterations. In this study, we used a first of its kind approach to target the Wwtr1-Camta1 gene fusion to the Wwtr1 locus, to develop a conditional EHE mouse model whereby Wwtr1-Camta1 is controlled by the endogenous transcriptional regulators upon Cre activation. These mice develop EHE tumors that are indistinguishable from human EHE clinically, histologically, immunohistochemically, and genetically. Overall, these results demonstrate unequivocally that TAZ-CAMTA1 is sufficient to drive EHE formation with exquisite specificity, as no other tumor types were observed. Furthermore, we fully credential this unique EHE mouse model as a valid preclinical model for understanding the role of TAZ dysregulation in cancer formation and for testing therapies directed at TAZ-CAMTA1, TAZ, and YAP/TAZ signaling.
Keywords: FLEx system; Hippo pathway; TAZ-CAMTA1; YAP/TAZ; endothelial cells; epithelioid hemangioendothelioma; fusion gene; mouse models of cancer; sarcoma.
© 2021 Seavey et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Similar articles
-
Unraveling the Biology of Epithelioid Hemangioendothelioma, a TAZ-CAMTA1 Fusion Driven Sarcoma.Cancers (Basel). 2022 Jun 16;14(12):2980. doi: 10.3390/cancers14122980. Cancers (Basel). 2022. PMID: 35740643 Free PMC article. Review.
-
WWTR1(TAZ)-CAMTA1 reprograms endothelial cells to drive epithelioid hemangioendothelioma.Genes Dev. 2021 Apr 1;35(7-8):495-511. doi: 10.1101/gad.348221.120. Epub 2021 Mar 25. Genes Dev. 2021. PMID: 33766984 Free PMC article.
-
Role of the Hippo-YAP/TAZ Pathway in Epithelioid Hemangioendothelioma and its Potential as a Therapeutic Target.Anticancer Res. 2024 Oct;44(10):4147-4153. doi: 10.21873/anticanres.17245. Anticancer Res. 2024. PMID: 39348982 Review.
-
TAZ-CAMTA1 and YAP-TFE3 alter the TAZ/YAP transcriptome by recruiting the ATAC histone acetyltransferase complex.Elife. 2021 Apr 29;10:e62857. doi: 10.7554/eLife.62857. Elife. 2021. PMID: 33913810 Free PMC article.
-
Loss of CDKN2A Cooperates with WWTR1(TAZ)-CAMTA1 Gene Fusion to Promote Tumor Progression in Epithelioid Hemangioendothelioma.Clin Cancer Res. 2023 Jul 5;29(13):2480-2493. doi: 10.1158/1078-0432.CCR-22-2497. Clin Cancer Res. 2023. PMID: 36598859
Cited by
-
Structure-based discovery of a novel small-molecule inhibitor of TEAD palmitoylation with anticancer activity.Front Oncol. 2022 Nov 29;12:1021823. doi: 10.3389/fonc.2022.1021823. eCollection 2022. Front Oncol. 2022. PMID: 36523977 Free PMC article.
-
YAP/TAZ as master regulators in cancer: modulation, function and therapeutic approaches.Nat Cancer. 2023 Jan;4(1):9-26. doi: 10.1038/s43018-022-00473-z. Epub 2022 Dec 23. Nat Cancer. 2023. PMID: 36564601 Free PMC article. Review.
-
The TAZ-CAMTA1 Fusion Protein Promotes Tumorigenesis via Connective Tissue Growth Factor and Ras-MAPK Signaling in Epithelioid Hemangioendothelioma.Clin Cancer Res. 2022 Jul 15;28(14):3116-3126. doi: 10.1158/1078-0432.CCR-22-0421. Clin Cancer Res. 2022. PMID: 35443056 Free PMC article. Clinical Trial.
-
LncRNA SFTA1P mediates positive feedback regulation of the Hippo-YAP/TAZ signaling pathway in non-small cell lung cancer.Cell Death Discov. 2021 Nov 29;7(1):369. doi: 10.1038/s41420-021-00761-0. Cell Death Discov. 2021. PMID: 34845189 Free PMC article.
-
Unraveling the Biology of Epithelioid Hemangioendothelioma, a TAZ-CAMTA1 Fusion Driven Sarcoma.Cancers (Basel). 2022 Jun 16;14(12):2980. doi: 10.3390/cancers14122980. Cancers (Basel). 2022. PMID: 35740643 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
