RAD21 is a driver of chromosome 8 gain in Ewing sarcoma to mitigate replication stress
- PMID: 33766983
- PMCID: PMC8015718
- DOI: 10.1101/gad.345454.120
RAD21 is a driver of chromosome 8 gain in Ewing sarcoma to mitigate replication stress
Abstract
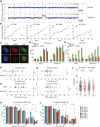
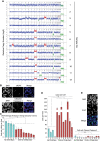
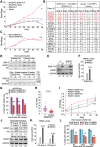
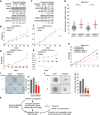
Aneuploidy, defined as whole-chromosome gain or loss, causes cellular stress but, paradoxically, is a frequent occurrence in cancers. Here, we investigate why ∼50% of Ewing sarcomas, driven by the EWS-FLI1 fusion oncogene, harbor chromosome 8 gains. Expression of the EWS-FLI1 fusion in primary cells causes replication stress that can result in cellular senescence. Using an evolution approach, we show that trisomy 8 mitigates EWS-FLI1-induced replication stress through gain of a copy of RAD21. Low-level ectopic expression of RAD21 is sufficient to dampen replication stress and improve proliferation in EWS-FLI1-expressing cells. Conversely, deleting one copy in trisomy 8 cells largely neutralizes the fitness benefit of chromosome 8 gain and reduces tumorgenicity of a Ewing sarcoma cancer cell line in soft agar assays. We propose that RAD21 promotes tumorigenesis through single gene copy gain. Such genes may explain some recurrent aneuploidies in cancer.
Keywords: DNA damage; Ewing sarcoma; RAD21; aneuploidy; cohesin; replication stress; trisomy 8.
© 2021 Su et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
