A novel lncRNA TCLlnc1 promotes peripheral T cell lymphoma progression through acting as a modular scaffold of HNRNPD and YBX1 complexes
- PMID: 33767152
- PMCID: PMC7994313
- DOI: 10.1038/s41419-021-03594-y
A novel lncRNA TCLlnc1 promotes peripheral T cell lymphoma progression through acting as a modular scaffold of HNRNPD and YBX1 complexes
Abstract
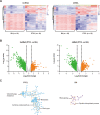
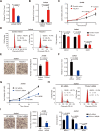
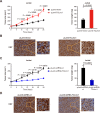
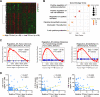
Long noncoding RNAs (lncRNAs) play an essential role in tumor progression. Few researches focused on the clinical and biological relevance of lncRNAs in peripheral T cell lymphoma (PTCL). In this research, a novel lncRNA (ENST00000503502) was identified overexpressed in the main subtypes of PTCL, and designated as T cell lymphoma-associated lncRNA1 (TCLlnc1). Serum TCLlnc1 was associated with extranodal involvement, high-risk International Prognostic Index, and poor prognosis of the patients. Both in vitro and in vivo, overexpression of TCLlnc1 promoted T-lymphoma cell proliferation and migration, both of which were counteracted by the knockdown of TCLlnc1 using small interfering RNAs. As the mechanism of action, TCLlnc1 directly interacted with transcription activator heterogeneous nuclear ribonucleoprotein D (HNRNPD) and Y-box binding protein-1 (YBX1) by acting as a modular scaffold. TCLlnc1/HNRNPD/YBX1 complex upregulated transcription of TGFB2 and TGFBR1 genes, activated the tumor growth factor-β signaling pathway, resulting in lymphoma progression, and might be a potential target in PTCL.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker.Biochim Biophys Acta Mol Basis Dis. 2017 Jul;1863(7):1805-1816. doi: 10.1016/j.bbadis.2017.04.014. Epub 2017 Apr 18. Biochim Biophys Acta Mol Basis Dis. 2017. PMID: 28428004
-
Long noncoding RNA PRKCQ-AS1 promotes CRC cell proliferation and migration via modulating miR-1287-5p/YBX1 axis.J Cell Biochem. 2020 Oct;121(10):4166-4175. doi: 10.1002/jcb.29712. Epub 2020 Jul 3. J Cell Biochem. 2020. PMID: 32619070
-
Long noncoding RNA SNHG6 promotes carcinogenesis by enhancing YBX1-mediated translation of HIF1α in clear cell renal cell carcinoma.FASEB J. 2021 Feb;35(2):e21160. doi: 10.1096/fj.202000732RR. Epub 2020 Nov 4. FASEB J. 2021. PMID: 33150667
-
LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network.Theranostics. 2020 Aug 29;10(23):10823-10837. doi: 10.7150/thno.47830. eCollection 2020. Theranostics. 2020. PMID: 32929382 Free PMC article.
-
Expression of cytotoxic proteins in peripheral T-cell and natural killer-cell (NK) lymphomas: association with extranodal site, NK or Tgammadelta phenotype, anaplastic morphology and CD30 expression.Leuk Lymphoma. 2000 Jul;38(3-4):317-26. doi: 10.3109/10428190009087022. Leuk Lymphoma. 2000. PMID: 10830738 Review.
Cited by
-
Epigenetic regulation in hematopoiesis and its implications in the targeted therapy of hematologic malignancies.Signal Transduct Target Ther. 2023 Feb 17;8(1):71. doi: 10.1038/s41392-023-01342-6. Signal Transduct Target Ther. 2023. PMID: 36797244 Free PMC article. Review.
-
RNA circuits and RNA-binding proteins in T cells.Trends Immunol. 2023 Oct;44(10):792-806. doi: 10.1016/j.it.2023.07.006. Epub 2023 Aug 18. Trends Immunol. 2023. PMID: 37599172 Free PMC article. Review.
-
Long non-coding RNA mitophagy and ALK-negative anaplastic lymphoma-associated transcript: a novel regulator of mitophagy in T-cell lymphoma.Haematologica. 2023 Dec 1;108(12):3333-3346. doi: 10.3324/haematol.2022.282552. Haematologica. 2023. PMID: 37381763 Free PMC article.
-
The role of lncRNA binding to RNA‑binding proteins to regulate mRNA stability in cancer progression and drug resistance mechanisms (Review).Oncol Rep. 2024 Nov;52(5):142. doi: 10.3892/or.2024.8801. Epub 2024 Sep 2. Oncol Rep. 2024. PMID: 39219266 Free PMC article. Review.
-
Circulating Long Non-Coding RNAs as Novel Potential Biomarkers for Osteogenic Sarcoma.Cancers (Basel). 2021 Aug 21;13(16):4214. doi: 10.3390/cancers13164214. Cancers (Basel). 2021. PMID: 34439367 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

