Immune memory in convalescent patients with asymptomatic or mild COVID-19
- PMID: 33767156
- PMCID: PMC7993859
- DOI: 10.1038/s41421-021-00250-9
Immune memory in convalescent patients with asymptomatic or mild COVID-19
Abstract
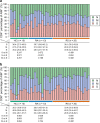
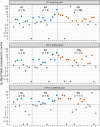
It is important to evaluate the durability of the protective immune response elicited by primary infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Here, we systematically evaluated the SARS-CoV-2-specific memory B cell and T cell responses in healthy controls and individuals recovered from asymptomatic or symptomatic infection approximately 6 months prior. Comparatively low frequencies of memory B cells specific for the receptor-binding domain (RBD) of spike glycoprotein (S) persisted in the peripheral blood of individuals who recovered from infection (median 0.62%, interquartile range 0.48-0.69). The SARS-CoV-2 RBD-specific memory B cell response was detected in 2 of 13 individuals who recovered from asymptomatic infection and 10 of 20 individuals who recovered from symptomatic infection. T cell responses induced by S, membrane (M), and nucleocapsid (N) peptide libraries from SARS-CoV-2 were observed in individuals recovered from coronavirus disease 2019 (COVID-19), and cross-reactive T cell responses to SARS-CoV-2 were also detected in healthy controls.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

