SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition
- PMID: 33767183
- PMCID: PMC7994801
- DOI: 10.1038/s41467-021-22166-4
SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition
Abstract
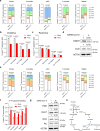
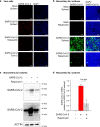
Viruses hijack host cell metabolism to acquire the building blocks required for replication. Understanding how SARS-CoV-2 alters host cell metabolism may lead to potential treatments for COVID-19. Here we profile metabolic changes conferred by SARS-CoV-2 infection in kidney epithelial cells and lung air-liquid interface (ALI) cultures, and show that SARS-CoV-2 infection increases glucose carbon entry into the TCA cycle via increased pyruvate carboxylase expression. SARS-CoV-2 also reduces oxidative glutamine metabolism while maintaining reductive carboxylation. Consistent with these changes, SARS-CoV-2 infection increases the activity of mTORC1 in cell lines and lung ALI cultures. Lastly, we show evidence of mTORC1 activation in COVID-19 patient lung tissue, and that mTORC1 inhibitors reduce viral replication in kidney epithelial cells and lung ALI cultures. Our results suggest that targeting mTORC1 may be a feasible treatment strategy for COVID-19 patients, although further studies are required to determine the mechanism of inhibition and potential efficacy in patients.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

