In silico studies reveal structural deviations of mutant profilin-1 and interaction with riluzole and edaravone in amyotrophic lateral sclerosis
- PMID: 33767237
- PMCID: PMC7994392
- DOI: 10.1038/s41598-021-86211-4
In silico studies reveal structural deviations of mutant profilin-1 and interaction with riluzole and edaravone in amyotrophic lateral sclerosis
Abstract
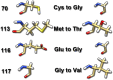
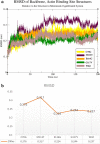
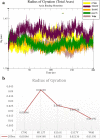
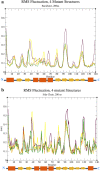
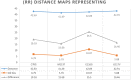
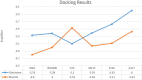
This study aimed to investigate four of the eight PFN-1 mutations that are located near the actin-binding domain and determine the structural changes due to each mutant and unravel how these mutations alter protein structural behavior. Swapaa's command in UCSF chimera for generating mutations, FTMAP were employed and the data was analyzed by RMSD, RMSF graphs, Rg, hydrogen bonding analysis, and RRdisMaps utilizing Autodock4 and GROMACS. The functional changes and virtual screening, structural dynamics, and chemical bonding behavior changes, molecular docking simulation with two current FDA-approved drugs for ALS were investigated. The highest reduction and increase in Rg were found to exist in the G117V and M113T mutants, respectively. The RMSF data consistently shows changes nearby to this site. The in silico data described indicate that each of the mutations is capable of altering the structure of PFN-1 in vivo. The potential effect of riluzole and edaravone two FDA approved drugs for ALS, impacting the structural deviations and stabilization of the mutant PFN-1 is evaluated using in silico tools. Overall, the analysis of data collected reveals structural changes of mutant PFN-1 protein that may explain the neurotoxicity and the reason(s) for possible loss and gain of function of PFN-1 in the neurotoxic model of ALS.
Conflict of interest statement
M.K. is the founder, president, and CEO of RockGen Therapeutics, LLC.
Figures



 ). On the actin side, residues are His 371, Glu 361, Tyr 169, Tyr 166, Ile 287, Asp 286, and 288. The amino acid residues involved in the hydrophobic interaction (
). On the actin side, residues are His 371, Glu 361, Tyr 169, Tyr 166, Ile 287, Asp 286, and 288. The amino acid residues involved in the hydrophobic interaction ( ) are identified and noted in the figure.
) are identified and noted in the figure.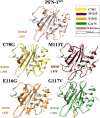

References
-
- Longo, D.L. Robert H. Brown, D. Phil., MD, and Ammar AlChalabi, Ph. D., FRCP, Dip. Stat. N. Engl. J. Med.377, 162–172 (2017).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

