Preliminary functional analysis of the subgingival microbiota of cats with periodontitis and feline chronic gingivostomatitis
- PMID: 33767308
- PMCID: PMC7994850
- DOI: 10.1038/s41598-021-86466-x
Preliminary functional analysis of the subgingival microbiota of cats with periodontitis and feline chronic gingivostomatitis
Abstract
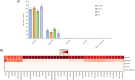
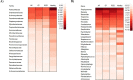
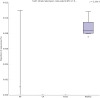
The subgingival microbial communities of domestic cats remain incompletely characterized and it is unknown whether their functional profiles are associated with disease. In this study, we used a shotgun metagenomic approach to explore the functional potential of subgingival microbial communities in client-owned cats, comparing findings between periodontally healthy cats and cats with naturally occurring chronic periodontitis, aggressive periodontitis, and feline chronic gingivostomatitis. Subgingival samples were subjected to shotgun sequencing and the metagenomic datasets were analyzed using the MG-RAST metagenomic analysis server and STAMP v2.1.3 (Statistical Analysis of Metagenomic Profiles) software. The microbial composition was also described to better understand the predicted features of the communities. The Respiration category in the level 1 Subsystems database varied significantly among groups. In this category, the abundance of V-Type ATP-synthase and Biogenesis of cytochrome c oxidases were significantly enriched in the diseased and in the healthy groups, respectively. Both features have been previously described in periodontal studies in people and are in consonance with the microbial composition of feline subgingival sites. In addition, the narH (nitrate reductase) gene frequency, identified using the KEGG Orthology database, was significantly increased in the healthy group. The results of this study provide preliminary functional insights of the microbial communities associated with periodontitis in domestic cats and suggest that the ATP-synthase and nitrate-nitrite-NO pathways may represent appropriate targets for the treatment of this common disease.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Feline chronic gingivostomatitis current concepts in clinical management.J Feline Med Surg. 2023 Aug;25(8):1098612X231186834. doi: 10.1177/1098612X231186834. J Feline Med Surg. 2023. PMID: 37548475 Free PMC article. Review.
-
The subgingival microbial community of feline periodontitis and gingivostomatitis: characterization and comparison between diseased and healthy cats.Sci Rep. 2019 Aug 26;9(1):12340. doi: 10.1038/s41598-019-48852-4. Sci Rep. 2019. PMID: 31451747 Free PMC article.
-
Case Report: Shift from Aggressive Periodontitis to Feline Chronic Gingivostomatitis Is Linked to Increased Microbial Diversity.Pathogens. 2025 Feb 26;14(3):228. doi: 10.3390/pathogens14030228. Pathogens. 2025. PMID: 40137713 Free PMC article.
-
Subgingival microbiota in individuals with severe chronic periodontitis.J Microbiol Immunol Infect. 2018 Apr;51(2):226-234. doi: 10.1016/j.jmii.2016.04.007. Epub 2016 May 13. J Microbiol Immunol Infect. 2018. PMID: 27262209
-
Subgingival microbial profile and production of proinflammatory cytokines in chronic periodontitis.Folia Med (Plovdiv). 2014 Jul-Sep;56(3):152-60. doi: 10.2478/folmed-2014-0022. Folia Med (Plovdiv). 2014. PMID: 25434071 Review.
Cited by
-
Feline Chronic Gingivostomatitis Diagnosis and Treatment through Transcriptomic Insights.Pathogens. 2024 Feb 21;13(3):192. doi: 10.3390/pathogens13030192. Pathogens. 2024. PMID: 38535535 Free PMC article.
-
Influence of Gallic Acid-Containing Mouth Spray on Dental Health and Oral Microbiota of Healthy Cats-A Pilot Study.Vet Sci. 2022 Jun 22;9(7):313. doi: 10.3390/vetsci9070313. Vet Sci. 2022. PMID: 35878330 Free PMC article.
-
Feline chronic gingivostomatitis current concepts in clinical management.J Feline Med Surg. 2023 Aug;25(8):1098612X231186834. doi: 10.1177/1098612X231186834. J Feline Med Surg. 2023. PMID: 37548475 Free PMC article. Review.
-
Epidemiological investigation of feline chronic gingivostomatitis and its relationship with oral microbiota in Xi'an, China.Front Vet Sci. 2024 Jun 14;11:1418101. doi: 10.3389/fvets.2024.1418101. eCollection 2024. Front Vet Sci. 2024. PMID: 38948672 Free PMC article.
-
Environmental risk factors for the development of oral squamous cell carcinoma in cats.J Vet Intern Med. 2022 Jul;36(4):1398-1408. doi: 10.1111/jvim.16372. Epub 2022 May 27. J Vet Intern Med. 2022. PMID: 35633064 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

