miR-23a/b suppress cGAS-mediated innate and autoimmunity
- PMID: 33767433
- PMCID: PMC8093233
- DOI: 10.1038/s41423-021-00668-x
miR-23a/b suppress cGAS-mediated innate and autoimmunity
Abstract
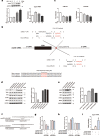
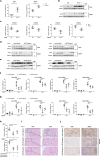
Cyclic GMP-AMP synthase (cGAS), a key sensor of intracellular DNA, is essential for eliciting innate immunity against infection, whereas aberrant activation of cGAS by endogenous DNA promotes severe autoimmune diseases. However, it is largely unknown how cGAS expression is regulated during pathogen infection and autoimmunity. Here, we report that during herpes simplex virus type 1 (HSV-1) infection, two microRNAs (miR-23a and miR-23b) whose levels significantly decrease due to their interaction with the lncRNA Oasl2-209 directly regulate the expression of cGAS. Overexpression of miR-23a/b markedly dampens cytosolic DNA-induced innate immune responses, whereas inhibition of miR-23a/b enhances these responses. Mice treated with miR-23a/b agomirs exhibit increased susceptibility to HSV-1 infection. Moreover, cGAS is significantly upregulated in the Trex1-/- mouse autoimmune disease model. Administration of miR-23a/b blunts self DNA-induced autoinflammatory responses in Trex1-/- mice. Collectively, our study not only reveals a novel regulatory mechanism of cGAS expression by miRNAs but also identifies a potential therapy for cGAS-related autoimmune diseases.
Keywords: Autoimmune disease; DNA virus; Innate immunity; cGAS; miR-23a; miR-23b.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

