Structural, Metabolic and Evolutionary Comparison of Bacterial Endospore and Exospore Formation
- PMID: 33767680
- PMCID: PMC7985256
- DOI: 10.3389/fmicb.2021.630573
Structural, Metabolic and Evolutionary Comparison of Bacterial Endospore and Exospore Formation
Abstract
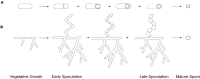
Sporulation is a specialized developmental program employed by a diverse set of bacteria which culminates in the formation of dormant cells displaying increased resilience to stressors. This represents a major survival strategy for bacteria facing harsh environmental conditions, including nutrient limitation, heat, desiccation, and exposure to antimicrobial compounds. Through dispersal to new environments via biotic or abiotic factors, sporulation provides a means for disseminating genetic material and promotes encounters with preferable environments thus promoting environmental selection. Several types of bacterial sporulation have been characterized, each involving numerous morphological changes regulated and performed by non-homologous pathways. Despite their likely independent evolutionary origins, all known modes of sporulation are typically triggered by limited nutrients and require extensive membrane and peptidoglycan remodeling. While distinct modes of sporulation have been observed in diverse species, two major types are at the forefront of understanding the role of sporulation in human health, and microbial population dynamics and survival. Here, we outline endospore and exospore formation by members of the phyla Firmicutes and Actinobacteria, respectively. Using recent advances in molecular and structural biology, we point to the regulatory, genetic, and morphological differences unique to endo- and exospore formation, discuss shared characteristics that contribute to the enhanced environmental survival of spores and, finally, cover the evolutionary aspects of sporulation that contribute to bacterial species diversification.
Keywords: Actinobacteria; Firmicutes; bacterial evolution; endospore; exospore; regulation; structural biology.
Copyright © 2021 Beskrovnaya, Sexton, Golmohammadzadeh, Hashimi and Tocheva.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Aldridge M., Facey P., Francis L., Bayliss S., Del Sol R., Dyson P. (2013). A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress? Nucleic Acids Res. 41 4813–4824. 10.1093/nar/gkt180 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

