Genome-Wide Identification of NAC Transcription Factors and Their Functional Prediction of Abiotic Stress Response in Peanut
- PMID: 33767732
- PMCID: PMC7985091
- DOI: 10.3389/fgene.2021.630292
Genome-Wide Identification of NAC Transcription Factors and Their Functional Prediction of Abiotic Stress Response in Peanut
Abstract
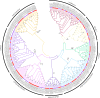
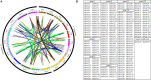
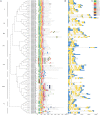
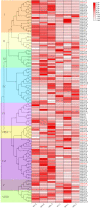
The NAC transcription factor (TF) is one of the most significant TFs in plants and is widely involved in plant growth, development, and responses to biotic and abiotic stresses. To date, there are no systematic studies on the NAC family in peanuts. Herein, 132 AhNACs were identified from the genome of cultivated peanut, and they were classified into eight subgroups (I-VIII) based on phylogenetic relationships with Arabidopsis NAC proteins and their conserved motifs. These genes were unevenly scattered on all 20 chromosomes, among which 116 pairs of fragment duplication events and 1 pair of tandem duplications existed. Transcriptome analysis showed that many AhNAC genes responded to drought and abscisic acid (ABA) stresses, especially most of the members in groups IV, VII, and VIII, which were expressed at larger differential levels under polyethylene glycol (PEG) and/or ABA treatment in roots or leaves. Furthermore, 20 of them selected in response to PEG and ABA treatment were evaluated by quantitative real-time polymerase chain reaction. The results showed that these genes significantly responded to drought and ABA in roots and/or leaves. This study was helpful for guiding the functional characterization and improvement of drought-resistant germplasms in peanuts.
Keywords: NAC transcription factor; abiotic stress; expression analysis; gene family; peanut.
Copyright © 2021 Li, Peng, Xu, Tang, Ma, Zhu, Shan and Wan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Diao W. P., John S., Wang S. B., Liu J. B., Pan B. G., Guo G. J., et al. (2018). Genome-wide analyses of the NAC transcription factor gene family in pepper (Capsicum annuum L.): chromosome location, phylogeny, structure, expression patterns, Cis-elements in the promoter, and interaction network. Int. J. Mol. Sci. 19 1048–1061. 10.3390/ijms19041028 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

