Extracellular cGAMP is a cancer cell-produced immunotransmitter involved in radiation-induced anti-cancer immunity
- PMID: 33768207
- PMCID: PMC7990037
- DOI: 10.1038/s43018-020-0028-4
Extracellular cGAMP is a cancer cell-produced immunotransmitter involved in radiation-induced anti-cancer immunity
Abstract
2'3'-cyclic GMP-AMP (cGAMP) is an intracellular second messenger that is synthesized in response to cytosolic double-stranded DNA and activates the innate immune STING pathway. Our previous discovery of its extracellular hydrolase ENPP1 hinted at the existence of extracellular cGAMP. Here, we detected that cGAMP is continuously exported but then efficiently cleared by ENPP1, explaining why it has previously escaped detection. By developing potent, specific, and cell impermeable ENPP1 inhibitors, we found that cancer cells continuously export cGAMP in culture at steady state and at higher levels when treated with ionizing radiation (IR). In mouse tumors, depletion of extracellular cGAMP decreased tumor-associated immune cell infiltration and abolished the curative effect of IR. Boosting extracellular cGAMP with ENPP1 inhibitors synergized with IR to delay tumor growth. In conclusion, extracellular cGAMP is an anti-cancer immunotransmitter that could be harnessed to treat cancers with low immunogenicity.
Conflict of interest statement
Competing Interests: M.S. and L.L. are scientific co-founders of Angarus Therapeutics, which has exclusive licensing rights to patent PCT/US2018/50018. J.A.C., V.B., K.E.S., M.S., and L.L. are inventors on patent PCT/US2018/50018.
Figures

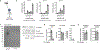





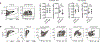


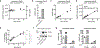
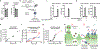




References
Methods References:
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

