Comparison of long-read sequencing technologies in interrogating bacteria and fly genomes
- PMID: 33768248
- PMCID: PMC8495745
- DOI: 10.1093/g3journal/jkab083
Comparison of long-read sequencing technologies in interrogating bacteria and fly genomes
Abstract
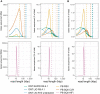
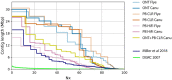
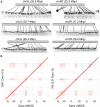
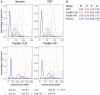
The newest generation of DNA sequencing technology is highlighted by the ability to generate sequence reads hundreds of kilobases in length. Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) have pioneered competitive long read platforms, with more recent work focused on improving sequencing throughput and per-base accuracy. We used whole-genome sequencing data produced by three PacBio protocols (Sequel II CLR, Sequel II HiFi, RS II) and two ONT protocols (Rapid Sequencing and Ligation Sequencing) to compare assemblies of the bacteria Escherichia coli and the fruit fly Drosophila ananassae. In both organisms tested, Sequel II assemblies had the highest consensus accuracy, even after accounting for differences in sequencing throughput. ONT and PacBio CLR had the longest reads sequenced compared to PacBio RS II and HiFi, and genome contiguity was highest when assembling these datasets. ONT Rapid Sequencing libraries had the fewest chimeric reads in addition to superior quantification of E. coli plasmids versus ligation-based libraries. The quality of assemblies can be enhanced by adopting hybrid approaches using Illumina libraries for bacterial genome assembly or polishing eukaryotic genome assemblies, and an ONT-Illumina hybrid approach would be more cost-effective for many users. Genome-wide DNA methylation could be detected using both technologies, however ONT libraries enabled the identification of a broader range of known E. coli methyltransferase recognition motifs in addition to undocumented D. ananassae motifs. The ideal choice of long read technology may depend on several factors including the question or hypothesis under examination. No single technology outperformed others in all metrics examined.
Keywords: Drosophila ananassae; bacterial genomics; fly genomics; genomics; sequencing.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
