Pathway testing for longitudinal metabolomics
- PMID: 33768548
- PMCID: PMC8252476
- DOI: 10.1002/sim.8957
Pathway testing for longitudinal metabolomics
Abstract
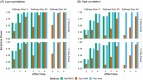
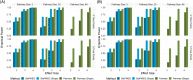
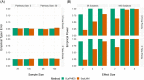
We propose a top-down approach for pathway analysis of longitudinal metabolite data. We apply a score test based on a shared latent process mixed model which can identify pathways with differentially progressing metabolites. The strength of our approach is that it can handle unbalanced designs, deals with potential missing values in the longitudinal markers, and gives valid results even with small sample sizes. Contrary to bottom-up approaches, correlations between metabolites are explicitly modeled leveraging power gains. For large pathway sizes, a computationally efficient solution is proposed based on pseudo-likelihood methodology. We demonstrate the advantages of the proposed method in identification of differentially expressed pathways through simulation studies. Finally, longitudinal metabolite data from a mice experiment is analyzed to demonstrate our methodology.
Keywords: global test; joint latent process; longitudinal analysis; mixed model; pseudo likelihood.
© 2021 The Authors. Statistics in Medicine published by John Wiley & Sons Ltd.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
