SARS-CoV-2 biology and variants: anticipation of viral evolution and what needs to be done
- PMID: 33769683
- PMCID: PMC8251359
- DOI: 10.1111/1462-2920.15487
SARS-CoV-2 biology and variants: anticipation of viral evolution and what needs to be done
Abstract
The global propagation of SARS-CoV-2 and the detection of a large number of variants, some of which have replaced the original clade to become dominant, underscores the fact that the virus is actively exploring its evolutionary space. The longer high levels of viral multiplication occur - permitted by high levels of transmission -, the more the virus can adapt to the human host and find ways to success. The third wave of the COVID-19 pandemic is starting in different parts of the world, emphasizing that transmission containment measures that are being imposed are not adequate. Part of the consideration in determining containment measures is the rationale that vaccination will soon stop transmission and allow a return to normality. However, vaccines themselves represent a selection pressure for evolution of vaccine-resistant variants, so the coupling of a policy of permitting high levels of transmission/virus multiplication during vaccine roll-out with the expectation that vaccines will deal with the pandemic, is unrealistic. In the absence of effective antivirals, it is not improbable that SARS-CoV-2 infection prophylaxis will involve an annual vaccination campaign against 'dominant' viral variants, similar to influenza prophylaxis. Living with COVID-19 will be an issue of SARS-CoV-2 variants and evolution. It is therefore crucial to understand how SARS-CoV-2 evolves and what constrains its evolution, in order to anticipate the variants that will emerge. Thus far, the focus has been on the receptor-binding spike protein, but the virus is complex, encoding 26 proteins which interact with a large number of host factors, so the possibilities for evolution are manifold and not predictable a priori. However, if we are to mount the best defence against COVID-19, we must mount it against the variants, and to do this, we must have knowledge about the evolutionary possibilities of the virus. In addition to the generic cellular interactions of the virus, there are extensive polymorphisms in humans (e.g. Lewis, HLA, etc.), some distributed within most or all populations, some restricted to specific ethnic populations and these variations pose additional opportunities for/constraints on viral evolution. We now have the wherewithal - viral genome sequencing, protein structure determination/modelling, protein interaction analysis - to functionally characterize viral variants, but access to comprehensive genome data is extremely uneven. Yet, to develop an understanding of the impacts of such evolution on transmission and disease, we must link it to transmission (viral epidemiology) and disease data (patient clinical data), and the population granularities of these. In this editorial, we explore key facets of viral biology and the influence of relevant aspects of human polymorphisms, human behaviour, geography and climate and, based on this, derive a series of recommendations to monitor viral evolution and predict the types of variants that are likely to arise.
© 2021 Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures

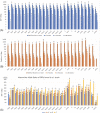

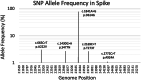
References
-
- Allen, C. , Bekoff, M. , and Lauder, G. (1998) Nature's Purposes. Cambridge, MA: MIT Press.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

