Maternal exposure to high-fat diet during pregnancy and lactation predisposes normal weight offspring mice to develop hepatic inflammation and insulin resistance
- PMID: 33769706
- PMCID: PMC7995551
- DOI: 10.14814/phy2.14811
Maternal exposure to high-fat diet during pregnancy and lactation predisposes normal weight offspring mice to develop hepatic inflammation and insulin resistance
Abstract
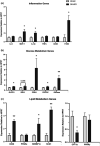
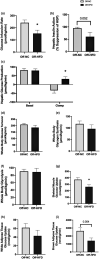
Increasing evidence shows a potential link between the perinatal nutrient environment and metabolic outcome in offspring. Here, we investigated the effects of maternal feeding of a high-fat diet (HFD) during the perinatal period on hepatic metabolism and inflammation in male offspring mice at weaning and in early adulthood. Female C57BL/6 J mice were fed HFD or normal chow (NC) for 4 weeks before mating and during pregnancy and lactation. The male offspring mice were weaned onto an NC diet, and metabolic and molecular experiments were performed in early adulthood. At postnatal day 21, male offspring mice from HFD-fed dams (Off-HFD) showed significant increases in whole body fat mass and fasting levels of glucose, insulin, and cholesterol compared to male offspring mice from NC-fed dams (Off-NC). The RT-qPCR analysis showed two- to fivefold increases in hepatic inflammatory markers (MCP-1, IL-1β, and F4/80) in Off-HFD mice. Hepatic expression of G6Pase and PEPCK was elevated by fivefold in the Off-HFD mice compared to the Off-NC mice. Hepatic expression of GLUT4, IRS-1, and PDK4, as well as lipid metabolic genes, CD36, SREBP1c, and SCD1 were increased in the Off-HFD mice compared to the Off-NC mice. In contrast, CPT1a mRNA levels were reduced by 60% in the Off-HFD mice. At postnatal day 70, despite comparable body weights to the Off-NC mice, Off-HFD mice developed hepatic inflammation with increased expression of MCP-1, CD68, F4/80, and CD36 compared to the Off-NC mice. Despite normal body weight, Off-HFD mice developed insulin resistance with defects in hepatic insulin action and insulin-stimulated glucose uptake in skeletal muscle and brown fat, and these metabolic effects were associated with hepatic inflammation in Off-HFD mice. Our findings indicate hidden, lasting effects of maternal exposure to HFD during pregnancy and lactation on metabolic homeostasis of normal weight offspring mice.
Keywords: high-fat diet; inflammation; insulin resistance; lactation; liver; maternal nutrition.
© 2021 The Authors. Physiological Reports published by Wiley Periodicals LLC on behalf of The Physiological Society and the American Physiological Society.
Conflict of interest statement
The authors have no conflicts to disclose.
Figures


References
-
- Ainge, H. , Thompson, C. , Ozanne, S. E. , & Rooney, K. B. (2011). A systematic review on animal models of maternal high fat feeding and offspring glycaemic control. International Journal of Obesity, 35(3), 325–335. - PubMed
-
- Ashino, N. G. , Saito, K. N. , Souza, F. D. , Nakutz, F. S. , Roman, E. A. , Velloso, L. A. , Torsoni, A. S. , & Torsoni, M. A. (2012). Maternal high‐fat feeding through pregnancy and lactation predisposes mouse offspring to molecular insulin resistance and fatty liver. The Journal of Nutritional Biochemistry, 23(4), 341–348. - PubMed
-
- Barker, D. J. P. (2007). The origins of the developmental origins theory. Journal of Internal Medicine, 261(5), 412–417. - PubMed
-
- Benatti, R. O. , Melo, A. M. , Borges, F. O. , Ignacio‐Souza, L. M. , Simino, L. A. P. , Milanski, M. (2014). Maternal high‐fat diet consumption modulates hepatic lipid metabolism and microRNA‐122 (miR‐122) and microRNA‐370 (miR‐370) expression in offspring. British Journal of Nutrition, 111(12), 2112–2122. - PubMed
-
- Buckley, A. J. , Keserü, B. , Briody, J. , Thompson, M. , Ozanne, S. E. , & Thompson, C. H. (2005). Altered body composition and metabolism in the male offspring of high fat–fed rats. Metabolism, 54(4), 500–507. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

