Discovery of New Protein Targets of BPA Analogs and Derivatives Associated with Noncommunicable Diseases: A Virtual High-Throughput Screening
- PMID: 33769846
- PMCID: PMC7997610
- DOI: 10.1289/EHP7466
Discovery of New Protein Targets of BPA Analogs and Derivatives Associated with Noncommunicable Diseases: A Virtual High-Throughput Screening
Abstract
Background: Bisphenol A analogs and derivatives (BPs) have emerged as new contaminants with little or no information about their toxicity. These have been found in numerous everyday products, from thermal paper receipts to plastic containers, and measured in human samples.
Objectives: The objectives of this research were to identify in silico new protein targets of BPs associated with seven noncommunicable diseases (NCDs), and to study their protein-ligand interactions using computer-aided tools.
Methods: Fifty BPs were identified by a literature search and submitted to a virtual high-throughput screening (vHTS) with 328 proteins associated with NCDs. Protein-protein interactions between predicted targets were examined using STRING, and the protocol was validated in terms of binding site recognition and correlation between in silico affinities and in vitro data.
Results: According to the vHTS, several BPs may target proteins associated with NCDs, some of them with stronger affinities than bisphenol A (BPA). The best affinity score (the highest in silico affinity absolute value) was obtained after docking 4,4'-bis(N-carbamoyl-4-methylbenzensulfonamide)diphenylmethane (BTUM) on estradiol 17-beta-dehydrogenase 1 (). However, other molecules, such as bisphenol A bis(diphenyl phosphate) (BDP), bisphenol PH (BPPH), and Pergafast 201 also exhibited great affinities (top 10 affinity scores for each disease) with proteins related to NCDs.
Discussion: Molecules such as BTUM, BDP, BPPH, and Pergafast 201 could be targeting key signaling pathways related to NCDs. These BPs should be prioritized for in vitro and in vivo toxicity testing and to further assess their possible role in the development of these diseases. https://doi.org/10.1289/EHP7466.
Figures




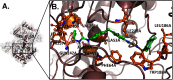
![Figure 5A is a three-dimensional view of the chemical complex structure of Bisphenol P H (B P P H) and Poly [A D P-ribose] polymerase 1 (P A R P 1) complex. Figure 5B is a three-dimensional view depicting the binding site and interactions for the following residues: T Y R 9 0 7 B, I L E 8 7 2 B, G L Y 8 6 3 B, A L A 8 9 8 B, T Y R 8 9 6 B, A L A 8 8 0 B, and T Y R 8 8 9 B. The arrows with circles depict hydrogen-bond donor features.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/351c/7997610/bf82e9328481/ehp7466_f5.gif)


References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. 2015. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25, 10.1016/j.softx.2015.06.001. - DOI
-
- Anderson BJ, inventor; 3M Innovative Properties Company, assignee. 2020. Resin blends of resorcinol diphthalonitrile ether with bisphenol M diphthalonitrile ether and/or bisphenol T diphthalonitrile ether. U.S. patent 10640598. 5 May 2020.
-
- Badrinarayanan P, Simon SL, Lyng RJ, O’Reilly JM. 2008. Effect of structure on enthalpy relaxation of polycarbonate: experiments and modeling. Polymer (Guildf) 49(16):3554–3560, 10.1016/j.polymer.2008.05.046. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
