A Comprehensive Map of mRNAs and Their Isoforms across All 14 Renal Tubule Segments of Mouse
- PMID: 33769951
- PMCID: PMC8017530
- DOI: 10.1681/ASN.2020101406
A Comprehensive Map of mRNAs and Their Isoforms across All 14 Renal Tubule Segments of Mouse
Abstract
Background: The repertoire of protein expression along the renal tubule depends both on regulation of transcription and regulation of alternative splicing that can generate multiple proteins from a single gene.
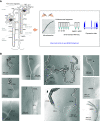
Methods: A full-length, small-sample RNA-seq protocol profiled transcriptomes for all 14 renal tubule segments microdissected from mouse kidneys.
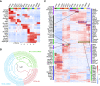
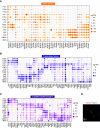
Results: This study identified >34,000 transcripts, including 3709 that were expressed in a segment-specific manner. All data are provided as an online resource (https://esbl.nhlbi.nih.gov/MRECA/Nephron/). Many of the genes expressed in unique patterns along the renal tubule were solute carriers, transcription factors, or G protein-coupled receptors that account for segment-specific function. Mapping the distribution of transcripts associated with Wnk-SPAK-PKA signaling, renin-angiotensin-aldosterone signaling, and cystic diseases of the kidney illustrated the applications of the online resource. The method allowed full-length mapping of RNA-seq reads, which facilitated comprehensive, unbiased characterization of alternative exon usage along the renal tubule, including known isoforms of Cldn10, Kcnj1 (ROMK), Slc12a1 (NKCC2), Wnk1, Stk39 (SPAK), and Slc14a2 (UT-A urea transporter). It also identified many novel isoforms with segment-specific distribution. These included variants associated with altered protein structure (Slc9a8, Khk, Tsc22d1, and Scoc), and variants that may affect untranslated, regulatory regions of transcripts (Pth1r, Pkar1a, and Dab2).
Conclusions: Full-length, unbiased sequencing of transcripts identified gene-expression patterns along the mouse renal tubule. The data, provided as an online resource, include both quantitative and qualitative differences in transcripts. Identification of alternative splicing along the renal tubule may prove critical to understanding renal physiology and pathophysiology.
Keywords: RNA-seq; alternative splicing; microdissection; renal tubule.
Copyright © 2021 by the American Society of Nephrology.
Figures





Comment in
-
Me Or Your Own Eyes: RNA-Seq and the Kidney.J Am Soc Nephrol. 2021 Apr;32(4):768-771. doi: 10.1681/ASN.2021010017. Epub 2021 Mar 4. J Am Soc Nephrol. 2021. PMID: 33769950 Free PMC article. No abstract available.
References
-
- Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z, et al.: Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol Cell 73: 130–142.e5, 2019 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

