One-step Reprogramming of Human Fibroblasts into Oligodendrocyte-like Cells by SOX10, OLIG2, and NKX6.2
- PMID: 33770499
- PMCID: PMC8072064
- DOI: 10.1016/j.stemcr.2021.03.001
One-step Reprogramming of Human Fibroblasts into Oligodendrocyte-like Cells by SOX10, OLIG2, and NKX6.2
Abstract
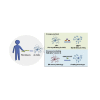
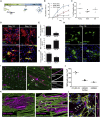
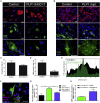
Limited access to human oligodendrocytes impairs better understanding of oligodendrocyte pathology in myelin diseases. Here, we describe a method to robustly convert human fibroblasts directly into oligodendrocyte-like cells (dc-hiOLs), which allows evaluation of remyelination-promoting compounds and disease modeling. Ectopic expression of SOX10, OLIG2, and NKX6.2 in human fibroblasts results in rapid generation of O4+ cells, which further differentiate into MBP+ mature oligodendrocyte-like cells within 16 days. dc-hiOLs undergo chromatin remodeling to express oligodendrocyte markers, ensheath axons, and nanofibers in vitro, respond to promyelination compound treatment, and recapitulate in vitro oligodendroglial pathologies associated with Pelizaeus-Merzbacher leukodystrophy related to PLP1 mutations. Furthermore, DNA methylome analysis provides evidence that the CpG methylation pattern significantly differs between dc-hiOLs derived from fibroblasts of young and old donors, indicating the maintenance of the source cells' "age." In summary, dc-hiOLs represent a reproducible technology that could contribute to personalized medicine in the field of myelin diseases.
Keywords: ATAC-seq; PMD; compound screenin; direct conversion; epigenetic age; human fibroblasts; oligodendrocytes.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Cailloux F., Gauthier-Barichard F., Mimault C., Isabelle V., Courtois V., Giraud G., Dastugue B., Boespflug-Tanguy O. Genotype-phenotype correlation in inherited brain myelination defects due to proteolipid protein gene mutations. Clinical European Network on Brain Dysmyelinating Disease. Eur. J. Hum. Genet. 2000;8:837–845. - PubMed
-
- Douvaras P., Fossati V. Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 2015;10:1143–1154. - PubMed
-
- Ehrlich M., Mozafari S., Glatza M., Starost L., Velychko S., Hallmann A.L., Cui Q.L., Schambach A., Kim K.P., Bachelin C. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc. Natl. Acad. Sci. U S A. 2017;114:E2243–E2252. - PMC - PubMed
-
- Fraga M.F., Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

