NaV1.2 EFL domain allosterically enhances Ca2+ binding to sites I and II of WT and pathogenic calmodulin mutants bound to the channel CTD
- PMID: 33770503
- PMCID: PMC8458505
- DOI: 10.1016/j.str.2021.03.002
NaV1.2 EFL domain allosterically enhances Ca2+ binding to sites I and II of WT and pathogenic calmodulin mutants bound to the channel CTD
Abstract
Neuronal voltage-gated sodium channel NaV1.2 C-terminal domain (CTD) binds calmodulin (CaM) constitutively at its IQ motif. A solution structure (6BUT) and other NMR evidence showed that the CaM N domain (CaMN) is structurally independent of the C-domain (CaMC) whether CaM is bound to the NaV1.2IQp (1,901-1,927) or NaV1.2CTD (1,777-1,937) with or without calcium. However, in the CaM + NaV1.2CTD complex, the Ca2+ affinity of CaMN was more favorable than in free CaM, while Ca2+ affinity for CaMC was weaker than in the CaM + NaV1.2IQp complex. The CTD EF-like (EFL) domain allosterically widened the energetic gap between CaM domains. Cardiomyopathy-associated CaM mutants (N53I(N54I), D95V(D96V), A102V(A103V), E104A(E105A), D129G(D130G), and F141L(F142L)) all bound the NaV1.2 IQ motif favorably under resting (apo) conditions and bound calcium normally at CaMN sites. However, only N53I and A102V bound calcium at CaMC sites at [Ca2+] < 100 μM. Thus, they are expected to respond like wild-type CaM to Ca2+ spikes in excitable cells.
Keywords: FRET; NMR; affinity; allostery; binding; biosensor; free energy; linkage; molecular recognition; titration; voltage-gated sodium channel.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

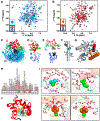


 , D24 ◪, K30 ▽, G33
, D24 ◪, K30 ▽, G33  , R37
, R37  , S38
, S38  , L39 ◧, G40
, L39 ◧, G40  , E45
, E45  , E54
, E54  G61 ◨, T62 ⊟, D64 ▷, F68
G61 ◨, T62 ⊟, D64 ▷, F68  , T70 ⧄, R74 ⊡. Inset: monitored CaMN residues (blue spheres) on 6BUT (gray). (D and E) Ca2+ titration of CaMC (sites III and IV) in 15N-CaM+14N-NaV1.2IQp. (D) Normalized peak intensities of apo CaM residues E82 ○, E83 ☒, F89 ⊡, V91 ☐, F92 ⧅, D93 △, K94
, T70 ⧄, R74 ⊡. Inset: monitored CaMN residues (blue spheres) on 6BUT (gray). (D and E) Ca2+ titration of CaMC (sites III and IV) in 15N-CaM+14N-NaV1.2IQp. (D) Normalized peak intensities of apo CaM residues E82 ○, E83 ☒, F89 ⊡, V91 ☐, F92 ⧅, D93 △, K94  , D95 ◪, Y99 ▽, S101
, D95 ◪, Y99 ▽, S101  , A102
, A102  , E104
, E104  , L105 ◧, H107
, L105 ◧, H107  , V108 ⧄, G113 ◨, E114 ⊟, K115
, V108 ⧄, G113 ◨, E114 ⊟, K115  , D122
, D122  , M124 ◩, I130 ⊞, Y138
, M124 ◩, I130 ⊞, Y138  , E139 ◩, E140 ⊲, T146 ⊳, A147 ✚, K148
, E139 ◩, E140 ⊲, T146 ⊳, A147 ✚, K148  . (E) Normalized peak intensities of (Ca2+)4-CaM residues E82 ○, E83 ☒, K94
. (E) Normalized peak intensities of (Ca2+)4-CaM residues E82 ○, E83 ☒, K94  , D95 ☐, Y99 ⧅, S101 △, A 102
, D95 ☐, Y99 ⧅, S101 △, A 102  , A103 ⧄, E104 ◪, G113 ⊞, E114 ▽, K115 ⊟, T117 ◩, R126
, A103 ⧄, E104 ◪, G113 ⊞, E114 ▽, K115 ⊟, T117 ◩, R126  , D129
, D129  , I130
, I130  , N137 ⊟, E139 ⊙, T146
, N137 ⊟, E139 ⊙, T146  , A147 ◫, K148 ◨. Inset: monitored CaMC residues (red spheres) on 6BUT (gray). (F–H) Ca2+ titration of 14N-CaM+15N-NaV1.2IQp. Normalized peak intensities of NaV1.2IQp residues. (F) K1901 ○, R1902 ◇, K1903 △, Q1904 ▽, E1905 ☐, E1906 ⧅, V1907
, A147 ◫, K148 ◨. Inset: monitored CaMC residues (red spheres) on 6BUT (gray). (F–H) Ca2+ titration of 14N-CaM+15N-NaV1.2IQp. Normalized peak intensities of NaV1.2IQp residues. (F) K1901 ○, R1902 ◇, K1903 △, Q1904 ▽, E1905 ☐, E1906 ⧅, V1907  , S1908 ☒, A1909
, S1908 ☒, A1909  , I1910 ⊟, I1911 ⧄; (G) I1912 ○, Q1913
, I1910 ⊟, I1911 ⧄; (G) I1912 ○, Q1913  , R1914 △, A1915 ☐, Y1916
, R1914 △, A1915 ☐, Y1916  , R1917
, R1917  , R1918 ⊞, Y1919
, R1918 ⊞, Y1919  , L1920 ⧅, L1921 ☒; and (H) Q1923 ◇, K1924 △, K1927
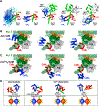
, L1920 ⧅, L1921 ☒; and (H) Q1923 ◇, K1924 △, K1927  . Insets: NaV1.2IQp residues monitored: pre-IQ/blue, IQ/green, post-IQ/red (spheres) on 6BUT (gray). Solid gray lines are for reference. (I and J) E1906/purple, A1909/black, I1912/cyan, Q1913/green, Y1916/orange, Y1919/orange (spheres) of NaV1.2IQp relative to apo (I) and (Ca2+)2-CaMC (J) surface (gray). 6BUT and 2M5E aligned by CaM 117–128 (blue spheres at 76). NaV1.2IQp 1,904–1,923 (gray helix), Cα of Q1904/blue and Q1923/red are small spheres. CaMC residues ≤4.5 Å of NaV1.2IQp with NaV1.2IQp-induced CSPs > (average plus 1 SD) are orchid (apo, I) or red ((Ca2+)4-CaM, J). See Figure S7.
. Insets: NaV1.2IQp residues monitored: pre-IQ/blue, IQ/green, post-IQ/red (spheres) on 6BUT (gray). Solid gray lines are for reference. (I and J) E1906/purple, A1909/black, I1912/cyan, Q1913/green, Y1916/orange, Y1919/orange (spheres) of NaV1.2IQp relative to apo (I) and (Ca2+)2-CaMC (J) surface (gray). 6BUT and 2M5E aligned by CaM 117–128 (blue spheres at 76). NaV1.2IQp 1,904–1,923 (gray helix), Cα of Q1904/blue and Q1923/red are small spheres. CaMC residues ≤4.5 Å of NaV1.2IQp with NaV1.2IQp-induced CSPs > (average plus 1 SD) are orchid (apo, I) or red ((Ca2+)4-CaM, J). See Figure S7.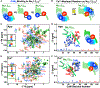


References
-
- Beaven GH, and Holiday ER (1952). Ultraviolet absorption spectra of proteins and amino acids. Adv. Protein Chem 7, 319–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

