A unified framework for cross-population trait prediction by leveraging the genetic correlation of polygenic traits
- PMID: 33770506
- PMCID: PMC8059341
- DOI: 10.1016/j.ajhg.2021.03.002
A unified framework for cross-population trait prediction by leveraging the genetic correlation of polygenic traits
Abstract
The development of polygenic risk scores (PRSs) has proved useful to stratify the general European population into different risk groups. However, PRSs are less accurate in non-European populations due to genetic differences across different populations. To improve the prediction accuracy in non-European populations, we propose a cross-population analysis framework for PRS construction with both individual-level (XPA) and summary-level (XPASS) GWAS data. By leveraging trans-ancestry genetic correlation, our methods can borrow information from the Biobank-scale European population data to improve risk prediction in the non-European populations. Our framework can also incorporate population-specific effects to further improve construction of PRS. With innovations in data structure and algorithm design, our methods provide a substantial saving in computational time and memory usage. Through comprehensive simulation studies, we show that our framework provides accurate, efficient, and robust PRS construction across a range of genetic architectures. In a Chinese cohort, our methods achieved 7.3%-198.0% accuracy gain for height and 19.5%-313.3% accuracy gain for body mass index (BMI) in terms of predictive R2 compared to existing PRS approaches. We also show that XPA and XPASS can achieve substantial improvement for construction of height PRSs in the African population, suggesting the generality of our framework across global populations.
Keywords: GWAS; UK Biobankcross-population; ancestry; cross-population; polygenic risk score.
Copyright © 2021 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




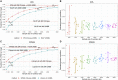
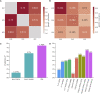

References
-
- Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018;19:581–590. - PubMed
-
- Abul-Husn N.S., Manickam K., Jones L.K., Wright E.A., Hartzel D.N., Gonzaga-Jauregui C., O’Dushlaine C., Leader J.B., Lester Kirchner H., Lindbuchler D.M. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354:aaf7000. - PubMed
-
- Craig J.E., Han X., Qassim A., Hassall M., Cooke Bailey J.N., Kinzy T.G., Khawaja A.P., An J., Marshall H., Gharahkhani P., NEIGHBORHOOD consortium. UK Biobank Eye and Vision Consortium Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat. Genet. 2020;52:160–166. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

