Meta-analysis of genome-wide DNA methylation identifies shared associations across neurodegenerative disorders
- PMID: 33771206
- PMCID: PMC8004462
- DOI: 10.1186/s13059-021-02275-5
Meta-analysis of genome-wide DNA methylation identifies shared associations across neurodegenerative disorders
Abstract
Background: People with neurodegenerative disorders show diverse clinical syndromes, genetic heterogeneity, and distinct brain pathological changes, but studies report overlap between these features. DNA methylation (DNAm) provides a way to explore this overlap and heterogeneity as it is determined by the combined effects of genetic variation and the environment. In this study, we aim to identify shared blood DNAm differences between controls and people with Alzheimer's disease, amyotrophic lateral sclerosis, and Parkinson's disease.
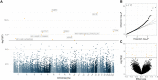
Results: We use a mixed-linear model method (MOMENT) that accounts for the effect of (un)known confounders, to test for the association of each DNAm site with each disorder. While only three probes are found to be genome-wide significant in each MOMENT association analysis of amyotrophic lateral sclerosis and Parkinson's disease (and none with Alzheimer's disease), a fixed-effects meta-analysis of the three disorders results in 12 genome-wide significant differentially methylated positions. Predicted immune cell-type proportions are disrupted across all neurodegenerative disorders. Protein inflammatory markers are correlated with profile sum-scores derived from disease-associated immune cell-type proportions in a healthy aging cohort. In contrast, they are not correlated with MOMENT DNAm-derived profile sum-scores, calculated using effect sizes of the 12 differentially methylated positions as weights.
Conclusions: We identify shared differentially methylated positions in whole blood between neurodegenerative disorders that point to shared pathogenic mechanisms. These shared differentially methylated positions may reflect causes or consequences of disease, but they are unlikely to reflect cell-type proportion differences.
Keywords: DNA methylation; Inflammatory markers; Methylation profile score; Mixed-linear models; Neurodegenerative disorders; Out-of-sample classification.
Conflict of interest statement
The authors declare that there are no competing interests.
Figures




References
-
- Pihlstrøm L, Wiethoff S, Houlden H. Chapter 22 - Genetics of neurodegenerative diseases: an overview. In: Kovacs GG, Alafuzoff I, editors. Handbook of Clinical Neurology. Volume 145: Elsevier; 2018. p. 309–23. 10.1016/b978-0-12-802395-2.00022-5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24 AG021886/AG/NIA NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- 108890/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- R01 AG033193/AG/NIA NIH HHS/United States
- 082604/2/07/Z/WT_/Wellcome Trust/United Kingdom
- TURNER/OCT15/972-797/MNDA_/Motor Neurone Disease Association/United Kingdom
- DH_/Department of Health/United Kingdom
- U01 AG016976/AG/NIA NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- 503480/MRC_/Medical Research Council/United Kingdom
- MR/R024804/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

