Defects in Emerin-Nucleoskeleton Binding Disrupt Nuclear Structure and Promote Breast Cancer Cell Motility and Metastasis
- PMID: 33771882
- PMCID: PMC8254762
- DOI: 10.1158/1541-7786.MCR-20-0413
Defects in Emerin-Nucleoskeleton Binding Disrupt Nuclear Structure and Promote Breast Cancer Cell Motility and Metastasis
Abstract
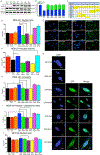
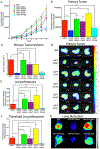
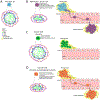
Nuclear envelope proteins play an important role in regulating nuclear size and structure in cancer. Altered expression of nuclear lamins are found in many cancers and its expression is correlated with better clinical outcomes. The nucleus is the largest organelle in the cell with a diameter between 10 and 20 μm. Nuclear size significantly impacts cell migration. Nuclear structural changes are predicted to impact cancer metastasis by regulating cancer cell migration. Here we show emerin regulates nuclear structure in invasive breast cancer cells to impact cancer metastasis. Invasive breast cancer cells had 40% to 50% less emerin than control cells, which resulted in decreased nuclear size. Overexpression of GFP-emerin in invasive breast cancer cells rescued nuclear size and inhibited migration through 3.0 and 8.0 μm pores. Mutational analysis showed emerin binding to nucleoskeletal proteins was important for its regulation of nuclear structure, migration, and invasion. Importantly, emerin expression inhibited lung metastasis by 91% in orthotopic mouse models of breast cancer. Emerin nucleoskeleton-binding mutants failed to inhibit metastasis. These results support a model whereby emerin binding to the nucleoskeleton regulates nuclear structure to impact metastasis. In this model, emerin plays a central role in metastatic transformation, because decreased emerin expression during transformation causes the nuclear structural defects required for increased cell migration, intravasation, and extravasation. IMPLICATIONS: Modulating emerin expression and function represents new targets for therapeutic interventions of metastasis, because increased emerin expression rescued cancer metastasis.
©2021 American Association for Cancer Research.
Conflict of interest statement
Figures
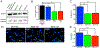



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

