Primate innate immune responses to bacterial and viral pathogens reveals an evolutionary trade-off between strength and specificity
- PMID: 33771921
- PMCID: PMC8020666
- DOI: 10.1073/pnas.2015855118
Primate innate immune responses to bacterial and viral pathogens reveals an evolutionary trade-off between strength and specificity
Abstract
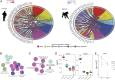
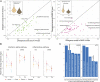
Despite their close genetic relatedness, apes and African and Asian monkeys (AAMs) differ in their susceptibility to severe bacterial and viral infections that are important causes of human disease. Such differences between humans and other primates are thought to be a result, at least in part, of interspecies differences in immune response to infection. However, because of the lack of comparative functional data across species, it remains unclear in what ways the immune systems of humans and other primates differ. Here, we report the whole-genome transcriptomic responses of ape species (human and chimpanzee) and AAMs (rhesus macaque and baboon) to bacterial and viral stimulation. We find stark differences in the responsiveness of these groups, with apes mounting a markedly stronger early transcriptional response to both viral and bacterial stimulation, altering the transcription of ∼40% more genes than AAMs. Additionally, we find that genes involved in the regulation of inflammatory and interferon responses show the most divergent early transcriptional responses across primates and that this divergence is attenuated over time. Finally, we find that relative to AAMs, apes engage a much less specific immune response to different classes of pathogens during the early hours of infection, up-regulating genes typical of anti-viral and anti-bacterial responses regardless of the nature of the stimulus. Overall, these findings suggest apes exhibit increased sensitivity to bacterial and viral immune stimulation, activating a broader array of defense molecules that may be beneficial for early pathogen killing at the potential cost of increased energy expenditure and tissue damage.
Keywords: early immune responses to infection; gram-negative bacteria; immunodeficiency viruses; pathogen-associated molecular patterns; primate evolution.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



Comment in
-
Innate immune responses between primates.Lab Anim (NY). 2021 May;50(5):121. doi: 10.1038/s41684-021-00769-0. Lab Anim (NY). 2021. PMID: 33911249 No abstract available.
References
-
- Redl H., Bahrami S., Schlag G., Traber D. L., Clinical detection of LPS and animal models of endotoxemia. Immunobiology 187, 330–345 (1993). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

