DNA replication origins retain mobile licensing proteins
- PMID: 33772005
- PMCID: PMC7998030
- DOI: 10.1038/s41467-021-22216-x
DNA replication origins retain mobile licensing proteins
Abstract
DNA replication in eukaryotes initiates at many origins distributed across each chromosome. Origins are bound by the origin recognition complex (ORC), which, with Cdc6 and Cdt1, recruits and loads the Mcm2-7 (MCM) helicase as an inactive double hexamer during G1 phase. The replisome assembles at the activated helicase in S phase. Although the outline of replisome assembly is understood, little is known about the dynamics of individual proteins on DNA and how these contribute to proper complex formation. Here we show, using single-molecule optical trapping and confocal microscopy, that yeast ORC is a mobile protein that diffuses rapidly along DNA. Origin recognition halts this search process. Recruitment of MCM molecules in an ORC- and Cdc6-dependent fashion results in slow-moving ORC-MCM intermediates and MCMs that rapidly scan the DNA. Following ATP hydrolysis, salt-stable loading of MCM single and double hexamers was seen, both of which exhibit salt-dependent mobility. Our results demonstrate that effective helicase loading relies on an interplay between protein diffusion and origin recognition, and suggest that MCM is stably loaded onto DNA in multiple forms.
Conflict of interest statement
The authors declare no competing interests.
Figures
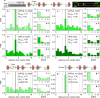
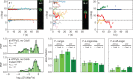
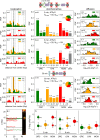

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

