Cargo sorting zones in the trans-Golgi network visualized by super-resolution confocal live imaging microscopy in plants
- PMID: 33772008
- PMCID: PMC7997971
- DOI: 10.1038/s41467-021-22267-0
Cargo sorting zones in the trans-Golgi network visualized by super-resolution confocal live imaging microscopy in plants
Abstract
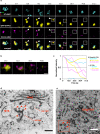
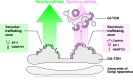
The trans-Golgi network (TGN) has been known as a key platform to sort and transport proteins to their final destinations in post-Golgi membrane trafficking. However, how the TGN sorts proteins with different destinies still remains elusive. Here, we examined 3D localization and 4D dynamics of TGN-localized proteins of Arabidopsis thaliana that are involved in either secretory or vacuolar trafficking from the TGN, by a multicolor high-speed and high-resolution spinning-disk confocal microscopy approach that we developed. We demonstrate that TGN-localized proteins exhibit spatially and temporally distinct distribution. VAMP721 (R-SNARE), AP (adaptor protein complex)-1, and clathrin which are involved in secretory trafficking compose an exclusive subregion, whereas VAMP727 (R-SNARE) and AP-4 involved in vacuolar trafficking compose another subregion on the same TGN. Based on these findings, we propose that the single TGN has at least two subregions, or "zones", responsible for distinct cargo sorting: the secretory-trafficking zone and the vacuolar-trafficking zone.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Staehelin LA, Moore I. The plant Golgi apparatus: structure, functional organization and trafficking mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995;46:261–288. doi: 10.1146/annurev.pp.46.060195.001401. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

