Host-derived lipids orchestrate pulmonary γδ T cell response to provide early protection against influenza virus infection
- PMID: 33772013
- PMCID: PMC7997921
- DOI: 10.1038/s41467-021-22242-9
Host-derived lipids orchestrate pulmonary γδ T cell response to provide early protection against influenza virus infection
Abstract
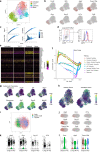
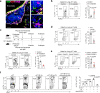
Innate immunity is important for host defense by eliciting rapid anti-viral responses and bridging adaptive immunity. Here, we show that endogenous lipids released from virus-infected host cells activate lung γδ T cells to produce interleukin 17 A (IL-17A) for early protection against H1N1 influenza infection. During infection, the lung γδ T cell pool is constantly supplemented by thymic output, with recent emigrants infiltrating into the lung parenchyma and airway to acquire tissue-resident feature. Single-cell studies identify IL-17A-producing γδ T (Tγδ17) cells with a phenotype of TCRγδhiCD3hiAQP3hiCXCR6hi in both infected mice and patients with pneumonia. Mechanistically, host cell-released lipids during viral infection are presented by lung infiltrating CD1d+ B-1a cells to activate IL-17A production in γδ T cells via γδTCR-mediated IRF4-dependent transcription. Reduced IL-17A production in γδ T cells is detected in mice either lacking B-1a cells or with ablated CD1d in B cells. Our findings identify a local host-immune crosstalk and define important cellular and molecular mediators for early innate defense against lung viral infection.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

