Social network analysis of the genealogy of strawberry: retracing the wild roots of heirloom and modern cultivars
- PMID: 33772307
- PMCID: PMC8022721
- DOI: 10.1093/g3journal/jkab015
Social network analysis of the genealogy of strawberry: retracing the wild roots of heirloom and modern cultivars
Erratum in
-
Corrigendum to: "Social network analysis of the genealogy of strawberry: retracing the wild roots of heirloom and modern cultivars".G3 (Bethesda). 2021 Sep 27;11(10):jkab257. doi: 10.1093/g3journal/jkab257. G3 (Bethesda). 2021. PMID: 34568936 Free PMC article. No abstract available.
Abstract
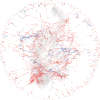
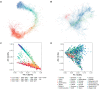
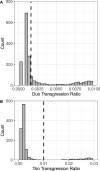
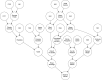
The widely recounted story of the origin of cultivated strawberry (Fragaria × ananassa) oversimplifies the complex interspecific hybrid ancestry of the highly admixed populations from which heirloom and modern cultivars have emerged. To develop deeper insights into the three-century-long domestication history of strawberry, we reconstructed the genealogy as deeply as possible-pedigree records were assembled for 8,851 individuals, including 2,656 cultivars developed since 1775. The parents of individuals with unverified or missing pedigree records were accurately identified by applying an exclusion analysis to array-genotyped single-nucleotide polymorphisms. We identified 187 wild octoploid and 1,171 F. × ananassa founders in the genealogy, from the earliest hybrids to modern cultivars. The pedigree networks for cultivated strawberry are exceedingly complex labyrinths of ancestral interconnections formed by diverse hybrid ancestry, directional selection, migration, admixture, bottlenecks, overlapping generations, and recurrent hybridization with common ancestors that have unequally contributed allelic diversity to heirloom and modern cultivars. Fifteen to 333 ancestors were predicted to have transmitted 90% of the alleles found in country-, region-, and continent-specific populations. Using parent-offspring edges in the global pedigree network, we found that selection cycle lengths over the past 200 years of breeding have been extraordinarily long (16.0-16.9 years/generation), but decreased to a present-day range of 6.0-10.0 years/generation. Our analyses uncovered conspicuous differences in the ancestry and structure of North American and European populations, and shed light on forces that have shaped phenotypic diversity in F. × ananassa.
Keywords: DNA forensics; Fragaria; biodiversity; conservation genetics; domestication; kinship.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures




References
-
- Affymetrix Inc. 2015. Axiom® Genotyping Solution Data Analysis Guide (P/N 702961 Rev. 3). Santa Clara, CA: Affymetrix, Inc..
-
- Ahmadi H, Bringhurst RS, Voth V.. 1990. Modes of inheritance of photoperiodism in Fragaria. J Am Soc Hortic Sci. 115:146–152.
-
- Barabási A-L. 2016. Network Science. Cambridge, UK: Cambridge University Press.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
